Biol/Chem 5310
Lecture: 12
October 3, 2002
Hemoglobin-2
Allostery
We have seen how CO2 and H+ affect oxygen bonding by Hb:
The binding of H+ and CO2 by Hb cause the release of oxygen from Hb.
This is known as allostery: The binding of ligand X by a protein at one site affects the binding of ligand Y at a different site on the same protein.
In this case, H+ and CO2 reduce the affinity of Hb for oxygen.
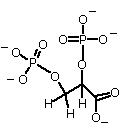
A third example of an allosteric effector is BPG:
2,3, Bisphosphoglycerate, a highly negatively charged molecule, synthesized in red blood cells.

Charge is about -4 at physiol. pH (Why?)
BPG decreases the affinity of Hb for oxygen by binding preferentially to deoxyHb
Some animals use other negatively-charged molecules in the same role: inositol hexaphosphate or ATP.
[BPG] increases at high altitude, where [O2] is lower. The decreased affinity of Hb for O2 allows a higher % of O2 to be released in the tissues
The effect of these allosteric effectors is to reduce the affinity of Hb for O2, thereby allowing more O2 to be released in the tissues, i.e. at low [O2]. How can we understand this in terms of protein structure?
First, the structures of Hb with and without O2 are different: called oxyHb and deoxyHb.
- Mb 153 aa
- Hb 141 aa a-subunit
- Hb 146 aa b-subunit
- a1-b1 interface contains ~35 aa (also a2-b2)
- This interface is the same in both conformations
- a1-b2 interface contains ~ 19 aa (also a2-b1)
- This interface is different in oxyHb and deoxyHb
- a1-a2 and b1-b2 interfaces involve only a few aa
How does O2 binding at one site in Hb initiate the conformational change?
O2 binding pulls the Fe2+ into the plane of the porphyrin ring Proximal His ligand is also pulled towards the heme Helix F, containing the proximal His, also moves Helix F translation causes a shift in positioning between a1 C helix and b2 FG region This change in the a1b2 interface causes all subunits to change from T to R See the Animation of Fig. 7-9
A series of salt bridges found in T-state Hb are broken during the transition to R-state {see structures}
Summary: The T-state (deoxyHb) is constrained by subunit interactions involving salt bridges and H-bonds to a conformation that binds O2 weakly-because of a long Fe2+-O2 bond. As more O2 binds, the energetically favorable Fe2+-O2 bonds form, eventually snapping the entire protein into the R-state (oxyHb)
Analysis of the Bohr effect Which protons are released when O2 binds? About 40% come from His146 at the C-terminus of b. The the T-state (deoxyHb) it forms a salt bridge with Asp94 of the same subunit. The salt bridge is broken in R-state (oxyHb) The His146 must be protonated in the salt bridge form: its pKa is 8.0 The pKa drops to 7.1 in the R-state form Since the pH is about 7.4, it will be mostly unprotonated. This accounts for some of the protons that are released upon O2 binding by Hb How does environment affect pKa values in proteins? A polar environment, especially oppositely charged as in an ion pair, favors the charged form of a group, e.g. COO- or NH3+ The pKa of a carboxyl group will be shifted down, favoring the unprotonated form The pKa value of an amino group will be shifted up, favoring the protonated form A nonpolar environment favors the uncharged form of a group, e.g. COOH or NH2 The pKa of a carboxyl group will be shifted up, favoring the protonated form The pKa value of an amino group will be shifted down, favoring the unprotonated form
Analysis of BPG binding
- N-teminus
- His2
- Lys82
- His143
- In this way BPG binding stabilizes the T state (deoxyHb)
- Fetal Hb has higher affinity for O2 than does adult (maternal) Hb.
- This promotes effective transfer of O2 from mother to fetus
- How to account for this?
- Fetal Hb is a2g2 with 2 gamma subunits instead of 2 beta subunits
- Gamma is very similar to beta, except that His143 is replaced by Ser
- This decreases the affinity of fetal Hb for BPG, and thereby increases its affinity for O2
Hemoglobin Variants
Because of the easy analysis of blood proteins many human variants (mutants) are known, with small deviations in the normal aa sequence. Some are much more common than others. Many are haarmless, some cause serious effects.
Examples:

Models for Cooperative Binding
A. Symmetry Model, MWC (Monod, Wyman, Changeux)
1. Oligomers of symmetrically related protomers
2. Each protomer has 2 conformations, R, T, that are in equilibrium
3. Ligand can bind to either conformation, but favors one
4. Symmetry is conserved during the conformational change: all subunits change conformation at once.
B. Sequential Model, based on Koshland's "induced fit" model for enzymes
In this model ligand binding can induce a conformational change in a protein. Subunits can change conformation, one at a time (sequentially). This model can explain negative cooperativity: if binding at one subunit causes a second subunit to change to a conformation with lower affinity.
C. Recent studies by Ackers indicate that a different set of rules apply for the conformational changes in Hb. Perutz, however, says the experiments are flawed (Annual Reviews of Biophysics, 1998).
Try a quiz in Chapter 7
Last updated
Comments/questions: svik@mail.smu.edu
Copyright 2002, Steven B. Vik, Southern Methodist University