Biol/Chem 5310
Lecture: 23
November 19, 2002
Nucleotides
Ch. 3 and Ch.23
Nucleic Acid Structure
Nucleotides are the building blocks of nucleic acids. Nucleic acids are linear polymers of nucleotides-with phosphodiester linkages. There are 2 types of nucleic acids:
RNA: ribonucleic acid
DNA: deoxyribonucleic acid
Roles: DNA is the genetic material of all cells and some viruses. Some viruses use RNA as genetic material. All cells use various types of RNA molecules for important functions. Examples: ATP is a single ribonucleotide.
large RNA's are used in protein synthesis by ribosomes.
Definitions:
Nucleotide is a phospho-ester of a nucleoside.
Nucleoside is an N-glycosidic linked "base" with a sugar (ribose or deoxyribose). Various examples:NADH, CoA, FAD
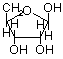
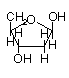
Ribose or deoxyribose are b-anomers
|
Ribose (b-D-ribofuranose)
|
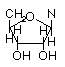
N-linked Ribose
|
 |
 |
|
Deoxyribose (b-D-deoxyribofuranose)
|
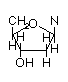
N-linked Deoxyribose
|
 |
 |
Primes signify sugar positions in nucleosides and nucleotides. Non-primed numbers signify base positions.
5 bases are found primarily in RNA and DNA (3 are common to both)
| Base | |
| Adenine | 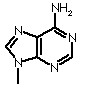 |
| Guanine | 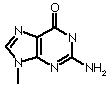 |
| Cytosine | 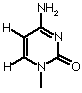 |
| Thymine | 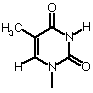 |
| Uracil | 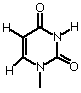 |
| Base | Single letter | Nucleoside | Classification |
| Adenine |
A |
Adenosine | Purine |
| Cytosine |
C |
Cytidine | Pyrimidine |
| Guanine |
G |
Guanosine | Purine |
| Thymine (DNA) |
T |
Thymidine | Pyrimidine |
| Uracil (RNA) |
U |
Uridine | Pyrimidine |
The nucleotides found in DNA and RNA are nucleoside-monophosphates. The phosphodiesters connect a phosphate to the 5 ' OH of one sugar to the 3 ' OH of another sugar.
Deoxyribose is 2 '-deoxy. Both ribose and deoxyribose have 3 ' and 5 ' OH groups.
So a linear nucleic acid has one free 5 ' OH at one end, and one 3 ' OH at the other end, just as a polypeptide has an amino terminus and a carboxy terminus, one at each end. By convention, the 5 ' end is written at the left, and the 3 ' end at the right.
A nucleic acid can be written:
pApCpGpT
or simply ACGT
Several Points:
| N6 methyl dA | 5 methyl dC |
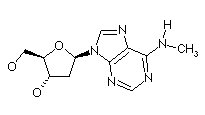 |
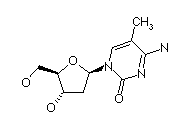 |
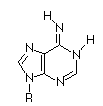
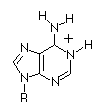
| Base |
Amino
|
Imino
|
|
Adenosine
|
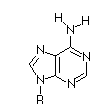 |
 |
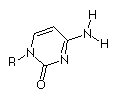
|
Cytosine
|
 |
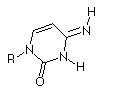 |
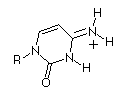
| Base |
Keto
|
Enol
|
|
Thymidine
|
 |
 |
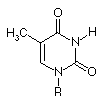
|
Guanosine
|
 |
 |
|
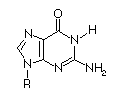
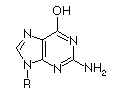
Base
|
Ionized
|
pKa
|
| Adenosine |
3.8
|
|
 |
 |
|
| Cytidine |
4.5
|
|
 |
 |
|
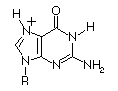
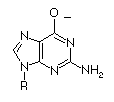
| Guanosine |
2.4
|
|
 |
 |
|
| Guanosine |
9.4
|
|
 |
How stable is DNA?
Base Pairing
Erwin Chargaff (1905-2002) (Bio) is credited with establishing that the base composition of DNA from any organism is fixed (for that organism) and that
% C =
% G % A =
% T 
At that time (~1940's) Hydrogen bonding was not seen as a key element of structure in molecular biology. Also, the predominant tautomers had not been established, so many possible pairings of the bases existed.
{To examine this problem see my Chime exercise.}
Even the X-ray diffraction data that Watson and Crick analyzed in the early 1950's did not reveal the hydrogen bonding of base-pairing. That is because the DNA analyzed was only a fiber of mixed composition DNA, not a crystal of a single molecular type. So it revealed only average structure.
Nevertheless, Watson and Crick were able to deduce, correctly, the structure of DNA in 1953.
It took another 25 years before true crystals of DNA (short pieces of a defined sequence) were solved by x-ray diffraction.
2 forms of DNA fibers were initially analyzed by Watson, Crick, Franklin, and Wilkins et al.Some of the samples were prepared by Rosalind Franklin, but she died of cancer in 1958 at the age of 37, before the Nobbel Prize was awarded.
| Rosalind Franklin |
The Nobel Prize in Medicine or Physiology was awarded to Watson, Crick & Wilkins in 1962 (Link)
 |
 |
 |
| James Watson | Francis Crick | Maurice Wilkins |
A-form (dehydrated)
B-form (hydrated)
B-form represents the structure generally found in living organisms.
Features of the B-form {see Chime link}
1) 2 polynucleotide strands are antiparallel, and wind around each other with a right-hand twist. End view is circular, with a 20 Å diameter.
2) Phosphates (and sugars) are on the outside-to minimize charge repulsion. Charge is further shielded by water and cations.
3) Bases are inside the structure, nearly perpendicular to the helix axis. Bases in one strand hydrogen bond to bases in the other strand, according to the rule: G to C, and A to T.
4) Dimensions: ~10 base pairs per turn, bases are ~3.4 Å thick and are closely packed (stacked). This yields a pitch of ~34 Å (rise per turn).
5) There are 2 surface grooves: Major and minor (by size). The ridges are formed by the deoxyribose-phosphates, and the bottoms of the grooves are formed by the edges of the bases.
2 additional points
1) the original model was made on average DNA. Now we have true crystals of short segments of DNA, 12 bp or 18 bp. {see link}. It is now known that details of the structure depend upon the actual DNA base sequence. e.g. TATATA looks different from GCGCGC. Proteins that bind to DNA can also distort the DNA.
2) The structure explains why %A=%T, and %G=%C in double-stranded DNA. Of all possible hydrogen bonding pairs, only these fit into this structure.
Other forms of DNA
1) A-form (dehydrated)
Bases are tilted ~20 degrees from the perpendicular to the helix axis. It is also a right-handed double helix. The pitch is ~28 Å., with 11 bp per turn. Major groove is shallow, and minor groove is deep. A-form DNA is similar to double-stranded RNA, e.g.transfer RNA., and RNA-DNA hybrids.
{see Chime link}
2) Z-form DNA
This is a left-handed double helix, that requires a repeating sequence of Pyrimidine-Purine-Pyrimidine-Purine......
Py = C or T, Pu = A or G
12 bp per turn, pitch = 45 Å.
Deep minor groove, with flat major groove
Purines are rotated relative to sugars (syn)
Pyrimidines are (anti) as in B or A-form DNA
Phosphates have a zig-zag pattern
{see Chime link}
Evidence indicates that short segments of Z-form exist in E.coli.
(See also the H-DNA)
Comparison of these forms of DNA {See Table 23-1}
| A form | B form | Z form | |
| Helical Sense | Right handed | Right handed | Left handed |
| Diameter | ~26 Å | ~20Å | ~18 Å |
| Base pairs per helical turn | 11 | 10 | 12 (6 dimers) |
| Helical twist per base pair | 33° | 36° | 60° (per dimer) |
| Helix pitch (rise per turn) | 28 Å | 34 Å | 45 Å |
| Helix rise per base pair | 2.6 Å | 3.4 Å | 3.7 Å |
| Base tilt normal to the axis | 20° | 6° | 7° |
| Major groove | Narrow & deep | Wide & deep | Flat |
| Minor groove | Wide & shallow | Narrow & deep | Narrow & deep |
| Sugar pucker | C3'-endo | C2'-endo | C2'-endo (pyrimidines), C3'-endo (purines) |
| Glycosidic bond | Anti | Anti | Anti (pyrimidines), syn (purines) |
See the Guided Exploration 21 on the CD-ROM for more images.
Last updated
Comments/questions: svik@mail.smu.edu
Copyright 2002, Steven B. Vik, Southern Methodist University