Biol/Chem 5310
Lecture: 25
November 25, 2002
Nucleic Acids
Structure of DNA and RNA forms, and DNA-Binding Proteins
Other forms of DNA
1) A-form (dehydrated)
Bases are tilted ~20 degrees from the perpendicular to the helix axis. It is also a right-handed double helix. The pitch is ~28 Å., with 11 bp per turn. Major groove is shallow, and minor groove is deep. A-form DNA is similar to double-stranded RNA, e.g.transfer RNA., and RNA-DNA hybrids.
{see Chime link}
2) Z-form DNA
This is a left-handed double helix, that requires a repeating sequence of Pyrimidine-Purine-Pyrimidine-Purine......
Py = C or T, Pu = A or G
12 bp per turn, pitch = 45 Å.
Deep minor groove, with flat major groove
Purines are rotated relative to sugars (syn)
Pyrimidines are (anti) as in B or A-form DNA
|
|
|
|
syn-Adenosine
|
anti-Adenosine
|
Phosphates have a zig-zag pattern
{see Chime link}
Evidence indicates that short segments of Z-form exist in E.coli.
(See also the H-DNA)
Comparison of these forms of DNA {See Table 23-1}
|
A form |
B form |
Z form |
| Helical Sense |
Right handed |
Right handed |
Left handed |
| Diameter |
~26 Å |
~20Å |
~18 Å |
| Base pairs per helical turn |
11 |
10 |
12 (6 dimers) |
| Helical twist per base pair |
33° |
36° |
60° (per dimer) |
| Helix pitch (rise per turn) |
28 Å |
34 Å |
45 Å |
| Helix rise per base pair |
2.6 Å |
3.4 Å |
3.7 Å |
| Base tilt normal to the axis |
20° |
6° |
7° |
| Major groove |
Narrow & deep |
Wide & deep |
Flat |
| Minor groove |
Wide & shallow |
Narrow & deep |
Narrow & deep |
| Sugar pucker |
C3'-endo |
C2'-endo |
C2'-endo (pyrimidines), C3'-endo (purines) |
| Glycosidic bond |
Anti |
Anti |
Anti (pyrimidines), syn (purines) |
See the Guided Exploration 21 on the CD-ROM for more images.
3) H-form DNA--an example of triple-helix nucleic acids
In the Chime example, 2 strands are parallel, and one is anti-parallel.
Two strands that are anti-parallel with respect to each other form somewhat normal H-bonding, while the third strand fits into the major groove, and also makes H-bonds via the bases.
Control of Supercoiling
Topoisomerases are enzymes that convert one topolological state or topoisomer to another. This is a change in L. These enzymes must break bonds in the strands of DNA, and then reform them.
2 examples:
1) Type I topoisomerases increase L by 1, by removing 1 negative supercoil. The enzyme makes a single-strand break (a nick), unwinds the helix one rotation, then reforms the second strand. (Chime link)
2) Type II procaryotic topoisomerases decrease L by 2 (DL = -2). They break both strands in a duplex. The reaction is driven by ATP because it is otherwise unfavorable. It increases negative supercoiling. Proposed mechanism in Fig. 23-14. (see Chime link)
Restriction Enzymes:
These are enzymes that cleave the strands of DNA at specific sites, according to the nucleotide sequence. They are counterparts of proteolytic enzymes such as trypsin, chymotrypsin in the world of nucleic acids. They are highly specific, and the sites they recognize may be very rare. Restriction enzymes are found in nature, primarily produced by bacteria as part of a defense system.
There are several types of restriction enzymes. We will focus on Type II, which is the type most widely used experimentally.
Nuclease: an enzyme that cleaves a nucleic acid
Restriction Endonuclease: an enzyme that cleaves within a segment of nucleic acid, when it recognizes a specific sequences of bases. (Restriction enzyme)
Typically, they are dimeric, and cleave at symmetrical sequences of DNA.
Examples EcoR I and BamH I
(See Chime link)
How do Restriction Enzymes recognize particular sequences of DNA?
Do they open up the strands, in order to see the bases?
No, they bind along the major or minor grooves, and interact with the edges of the bases, for sequences specificity,
and also with the sugars and the phosphates, for non-specific binding. This involves, H-bonding, hydrophobic contacts, and electrostatic interactions (ion pairs).
How is the expression of genes controlled?
1. Prokaryotic Repressors are proteins that bind to sequences of DNA (operators) and prevent transcription (RNA polymerase)
Motifs: helix-turn-helix, example Lambda repressor (Chime)
anti-parallel beta strand, example Met repressor
2. Eukaryotic transcriptional activators
1. GAL4 : Dimeric, coiled coil--contacts DNA via Zn-binding domains (Chime)
2. GCN4: Dimeric, coiled coil--contacts DNA vis basic alpha-helix (Chime)
Organization of Eukaryotic DNA:
The Nucleosome: about 150 bp of DNA with 8 proteins, 2 each of the Histones 2A, 2B, 3, and 4.
These are basic proteins that stabilize the negative charge of DNA. In particular there are Arginine residues that fit into the minor groove. (Chime)
RNA structure:
1. tRNA: about 76 nucleotides, folded into stems and loops with extensive base-pairing and base-stacking to form a compact "L"-shaped structure. The 3' end carries an amino acid to the ribosome for protein synthesis. (Chime)
Base include non-standard ones
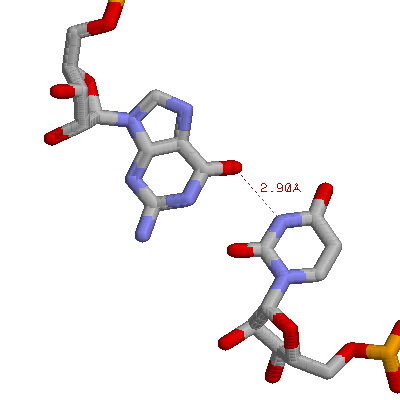
There is non-standard base-pairing. Shown above is a G:U base pair from phenylalanine tRNA (G4 and 69U)
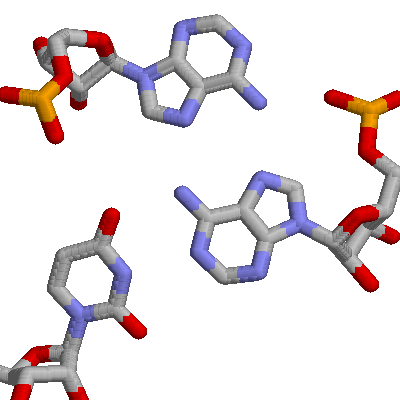
There are tertiary interactions, involving 3 bases. Shown below is an A:U base pair, along with a second A that H-bonds to the first A. These are A9, U12, and A23 from the yeast phenylalanine tRNA.
2. The Ribosome. This is an enormous structure that is the site of protein synthesis. It contains two subunits. The smaller one is shown in Chime.
| Ribosome from E. coli |
|
|
| Large Subunit |
31 Proteins (30%) |
2 RNA, 2904 and 120 nucleotides |
| Small Subunit |
21 proteins (40%) |
1 RNA, 1542 nucleotides |
Last updated Monday, November 25, 2002
Comments/questions: svik@mail.smu.edu
Copyright 2002, Steven B. Vik, Southern Methodist University