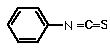Biol/Chem 5310
Lecture: 7
September 12, 2002
Covalent Structures of Proteins
Proteins are formed when amino acids are linked by peptide bonds.
Proteins are linear polymers (polypeptides) of amino acids.
In proteins, amino acids are often called amino acid residues.
There are 4 levels of organization in protein structure:
- Primary: amino acid sequence
- Secondary: patterns of backbone conformation, e.g. a-helix, b-sheet, turns
- Tertiary: the arrangement of the elements of secondary structure in space, or the 3-D structure
- Quaternary: the arrangement of individual subunits in a multi-subunit protein
Today focus on primary structure.
- To understand the function of a protein it is necessary to know its primary structure, or sequence.
- By convention the sequence of polypeptides is always written, from left to right, N-terminus to C-terminus.
- The N-terminus refers to the end of the chain with the free a-amino group. There can be only one, although sometimes it is covalently modified (blocked).
- The C-terminus refers to the other end, which has a free carboxy group. This too can be covalently modified.
How to determine the sequence of a protein?
To see the amino acid structures , check the link: Structures
- Often one determines the sequence of the cloned gene, and translates it by computer to the protein sequence
- Sometimes the gene or the initial protein is modified, so that it is necessary to analyze the mature protein itself.
- In the past it was likely that the cloned gene was not available, so it was necessary to work with the protein.
1) End group analysis
N-terminus
This method was used by Fred Sanger to determine the sequence of insulin, for which he was awarded the Nobel Prize in Chemistry in 1958
 link to the Nobel site, historical link
Determines the amino acid at the N-terminus
Can reveal if there are multiple polypeptides in the protein, or sample
Dansyl chloride
link to the Nobel site, historical link
Determines the amino acid at the N-terminus
Can reveal if there are multiple polypeptides in the protein, or sample
Dansyl chloride
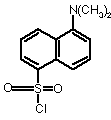 reacts with free amino groups to make a fluorescent product. After acid hydrolysis to produce free amino acids, they are resolved by chromatography, and detected by fluorescence.
Edman reagent, (See Animation of Fig. 5-15)
reacts with free amino groups to make a fluorescent product. After acid hydrolysis to produce free amino acids, they are resolved by chromatography, and detected by fluorescence.
Edman reagent, (See Animation of Fig. 5-15)
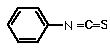 phenyl isothiocyante (PITC) also reacts with amino groups. This procedure can be done repetitively, as the covalently modified N-terminal amino acid can be selectively released.
The Edman degradation has also been automated, using "sequenators" that can carry out 50 or more cycles, thereby determining a significant portion of the N-terminal sequence
Pehr Victor Edman, courtesy of the Australian Academy of Science
phenyl isothiocyante (PITC) also reacts with amino groups. This procedure can be done repetitively, as the covalently modified N-terminal amino acid can be selectively released.
The Edman degradation has also been automated, using "sequenators" that can carry out 50 or more cycles, thereby determining a significant portion of the N-terminal sequence
Pehr Victor Edman, courtesy of the Australian Academy of Science

C-terminus
- Chemistry has been developed recently for an Edman-like degradation of C-terminal residues, but it is not widely used yet.
- Traditionally, enzymes have been used to release C-terminal residues. These enzymes work repetitively and continuously, but can be slowed for detection of the first few residues.
- These enzymes, carboxypeptidases, have different specificities.
- Carboxypeptidase A cannot release Arg, Lys, or Pro at the terminal position. Also, no amino acid will be released if Pro is at the penultimate position.
- Carboxypeptidase B will release only Lys or Arg at the terminal position, unless Pro is at the penultimate position.
- Carboxypeptidase C can release any amino acid.
- Carboxypeptidase Y can also release any amino acid, but Gly is very slow.
2) Separate chains linked by Disulfide bonds
- Oxidation to cysteic acid (-SO3-) by performic acid (HCOOOH)
-
- Reduction to sulfhydryl (-SH) by 2-mercaptoethanol or DTT (dithiothreitol). This is usually followed by alkylation for stability: using iodoacetate (ICH2COO-)
3) Amino Acid Composition
- Complete hydrolysis to indiviual amino acids, followed by chromatography for identification.
- Acid hydrolysis of peptide bonds
- 6 M HCl, 100 °C, 10-100 hrs
- Trp is destroyed
- Asn, Gln are deamidated to Asp, Glu so Asx and Glx are determined
- Base hydrolysis is used to determine Trp
- Analysis of the hydrolyzed amino acids is usually by HPLC (High Pressure Liquid Chromatography)
4) If the polypeptide is too long to be sequenced completely by repetive Edman degradation (the usual case), it must be fragmented.
The fragment must be sequence specific for long proteins. Two different methods are used: chemical treatments and proteolytic enzymes. (See the Animation of Fig. 5-16)
Enzymes
- Trypsin is a digestive enzyme that cleaves proteins at peptide bonds that are on the C-side of a Lys or Arg
- Chymotrypsin is a similar enzyme that cleaves on the C-side of Trp, Tyr, Phe (and sometimes others)
- Endopeptidase V8 cleaves on the C-side of Glu
- Elastase, Thermolysin, and Pepsin have broader specificites
Chemical fragmentation
-
Br-CN (Cyanogen Bromide) is the most commonly used.
It attacks Met, fragmenting the polypeptide on the C-side of the Met, and altering the Met to homoserine lactone
Example:
For the following peptide Ala-Gly-Arg-Ser-Met-Phe
These results can be used to determine the sequence:
- N-terminal analysis: Ala Result
- Trypsin digestion reveals 2 fragments with the following compositions:
- (Ala, Arg, Gly)
- (Met, Phe, Ser)
- Cyanogen Bromide treatment yields 2 fragments:
- (Ala, Arg, Gly, Met*, Ser)
- (Phe)
Guided Exploration #4 from the CD. This is a very good guide to protein sequence analysis.
Try a quiz in Ch. 5
Last updated Friday, September 6, 2002
Comments/questions: svik@mail.smu.edu
Copyright 2002, Steven B. Vik, Southern Methodist University
link to the Nobel site, historical link