 |
 |
 |
This package contains a series of boundary integral Poisson-Boltzmann (PB) solvers. Boundary integral PB solvers is a smart choice if the accuracy of electrostatic potentials and fields near or on the molecular surface are the major concerns of one's research. Among these solvers, TABI-PB generally solves the PB equation with the best combination of efficiency and accuracy. HOBI-PB provides the most accurate electrostatic potential on the triangular molecular surface at the cost of increased CPU time and Memory use. GABI-PB performs extremely well for solving PB equation with less than 300,000 boundary elements as it utilizes the GPU-accelerations under the direct summation scheme (no treecode). Interested users can download these solvers and use them for academic purpose by following the New BSD License and citing the related publication as specified below.
This material is based upon work supported by the National Science Foundation under NSF Grant DMS-0915057, DMS-1418966, DMS-1418957, DMS-1819094, DMS-1819193. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
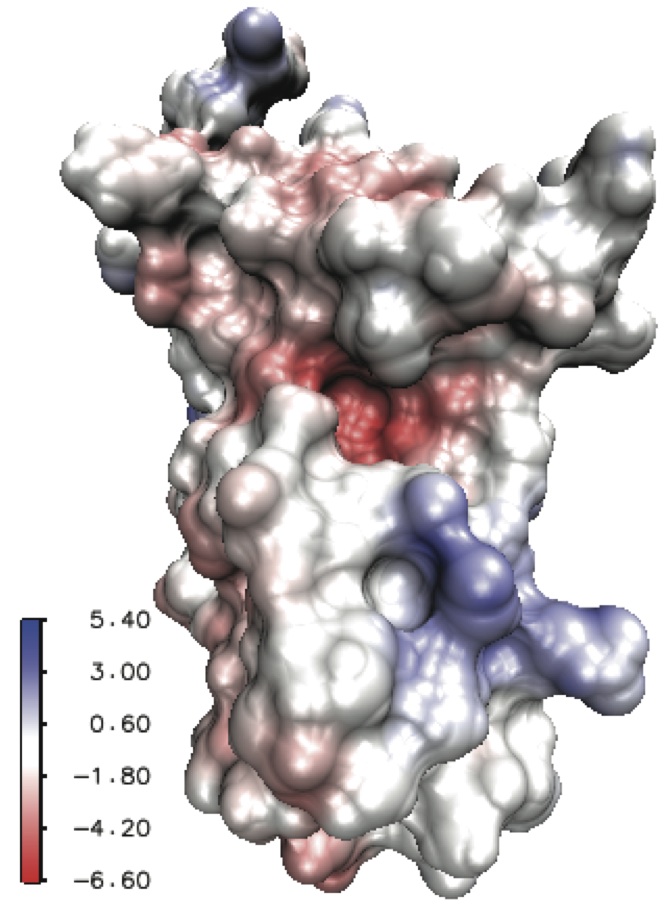
This software package employs a well-conditioned boundary integral formulation for the electrostatic potential and its normal derivative on the molecular surface. The surface is triangulated and the integral equations are discretized by centroid collocation. The linear system is solved by GMRES iteration and the matrix-vector product is carried out by a Cartesian terraced which reduces the cost from O(N^2) to O(N*logN), where N is the number of faces in the triangulation. The TABI solver can be applied to compute the electrostatic potential on molecular surface and solvation energy. The solver can be obtained with two different approaches.
One way is to down the binary version package (Windows, Linux, Mac). Note: For this option, testing proteins in forms of APBS's pqr file (sample) should be stored in the sub-directory with the name of "test_proteins", while MSMS of the corresponding operation system (Windows, Linux, Mac), and the input file (sample) should be in the same directory with the binary files. You may also need to change the property of the binary files (tabipb.exe and msms) to executable. More details can be found from the User's Guide.
The other way is to download the sourcecode (written in Fortran 77/90/95) from sourceforge and more details can be found from the User's Guide.
REFERENCE: W.H. Geng and R. Krasny, A treecode-accelerated boundary integral Poisson-Boltzmann solver for continuum electrostatics of solvated biomolecules, J. Comput. Phys. 247, 62-87 (2013).
This software contains the parallel higher-order boundary integral method to solve the linear Poisson-Boltzmann (PB) equation. In our method, a well-posed boundary integral formulation is used to ensure the fast convergence of Krylov subspace linear solver such as GMRES. The molecular surfaces are first discretized with flat triangles and then converted to curved riangles with the assistance of normal information at vertices. To maintain the desired accuracy, four-point Gauss-Radau quadratures are used on regular triangles and sixteen-point Gauss-Legendre quadratures together with regularization transformations are applied on singular triangles. We take advantage of the embarrassingly parallel feature of boundary integral formulation, and parallelize the schemes with the message passing interface (MPI) implementation. .
Users can download the source code (written in Fortran 77/90/95 with Open MPI) from sourceforge. Note the Users need to have Open MPI installed on ones' system.
REFERENCE: W.H. Geng, Parallel higher-order boundary integral electrostatics computation on molecular surfaces with curved triangulation, J. Comput. Phys., 241, 253-265 (2013).
This software package presents a GPU-accelerated direct-sum boundary integral method to solve the linear Poisson-Boltzmann (PB) equation. In our method, a well-posed boundary integral formulation is used to ensure the fast convergence of Krylov subspace based linear algebraic solver such as the GMRES. The molecular surfaces are discretized with flat triangles and centroid collocation. To speed up our method, we take advantage of the parallel nature of the boundary integral formulation and parallelize the schemes within CUDA shared memory architecture on GPU. The schemes use only $11N+6N_c$ size-of-double device memory for a biomolecule with $N$ triangular surface elements and $N_c$ partial charges.
Users can download the source code (written in CUDA C) from sourceforge. Note the Users need to have GPUs access and nvcc complier enabled.
REFERENCE: W.H. Geng and F. Jacob, A GPU-accelerated direct-sum boundary integral Poisson-Boltzmann solver, Comput. Phys. Commun. 184, 1490-1496 (2013).