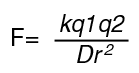
Biol/Chem 5310
Lecture: 3
August 29, 2002
Noncovalent Interactions/pH Calculations
Noncovalent Interactions are discussed on pp. 25-26 of your text, and in Table 2-1
These interactions can be organized in several different ways. For example:
- dipoles, induced dipoles and dispersion forces
Charge-Charge Interactions
The Force between 2 point charges is determined by Coulomb's Law
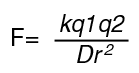
where q1 and q2 are the charges, r is the distance between the charges and D is the Dielectric constant of the medium.
In biochemistry we are usually interested in the Energy:

In water D is about 80, whereas in organic solvents D is about 2-10. In a vacuum, D=1. Therefore in water ionic interactions are diminished in strength. However, the inside of a protein more closely resembles an organic solvent, in terms of an "effective dielectric constant", so these energies can be greater than in water. The equation above tells us that the energy is a spherically-symmetrical function, i.e. it is not directional. Also, the energy falls off as 1/r , which means that although it declines with distance, it does so only gradually.
Dipoles:
A dipole is a molecule with an uneven distribution of charge. It has a positive and a negative end. These could be charges or partial charges. The dipole moment is equal to the charge separated times the distance of separation.
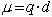
Dipole moments:
| Water | 1.83 Debye | |
| Ammonia | 1.48 Debye | |
| Glycine | 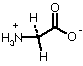 |
16.7 Debye |
O=C=O has a zero dipole moment
Dipolar interactions are directional, according to the charge distributon. For example the negative end of water will interact more favorably with a positive ion. Dipolar interactions fall off more quickly with distance than do charge-charge interactions . So for an interaction of 2 dipoles, the energy goes as 1/r3.
Dispersion Forces (or simply called van der waals interactions). All molecules have a capacity for weak attraction at close range, but with high repulsion at even closer range. This leads to a vanderwaals radius for nonbonded contact.
Hydrogen Bonds:
This is a type of electrostatic interaction involving a partially positive Hydrogen atom and a partially negative acceptor atom, that is usually Oxygen, and sometimes Sulfur or Nitrogen. Hydrogen bonds are very important in the specificity of DNA interactions in forming duplexes, and in the formation of alpha-helices in protein structures. The strength of H-bonds depends upon both distance and orientation of the donors and acceptors.
1) Calculate the pH of 0.1 M Acetic Acid
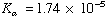
pKa = 4.76 Solve by approximating [HA] as 0.1 M (very little dissociation), then check 
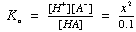

Or solve "exactly" using the quadratic equation 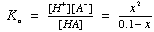
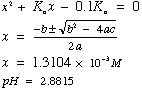
Approximation works if [HA]initial >> Ka
2) Calculate the pH of 0.1 M ammonia
pKa = 9.25
ammonia is a weak base: 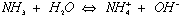
approximate by assuming [NH3} remains about 0.1M 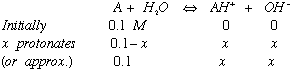
solve for x, convert to pOH, then calculate pH 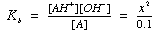
You can check your answers with the linked spreadsheet.
3) Polyprotic acids: phosphoric acid
3 pKa's: 2.15, 6.82, 12.38 4 ionic species with charges of 0, -1, -2, and -3 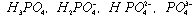
pH of is as in part 1) using pKa1 = 2.15 (only consider the first dissociation)
pH of is as in part 2) using pKa3 = 12.38 (only consider the first protonation of PO4-3)
pH of is 1/2 (pKa1 + pKa2) = 4.485 (the average of the protonation and deprotonation steps)
pH of is 1/2 (pKa2 + pKa3) =9.60 (the average of the protonation and deprotonation steps)
Another problem: What are the concentrations of all species at pH=7.2 (0.1 M)?
Since pKa1 << 7.2 and pKa3 >> 7.2 We can guess that only and
are significant
Because remember the HH eqn: the ratio of is pH - pKa1 = 5.05
or about 100,000 times more than

Use the HH eqn to calculate the amount of each species.
Set up
Try the Chapter 2 quizUse 3 HH eqn's. one for each pK Label species A, B, C, and D A is H3PO4, B is H2PO4- etc A + B + C + D = 0.1M Four eqn's, four unknowns
Back to 5310 Home
Last updated
Comments/questions: svik@mail.smu.edu
Copyright 2002, Steven B. Vik, Southern Methodist University