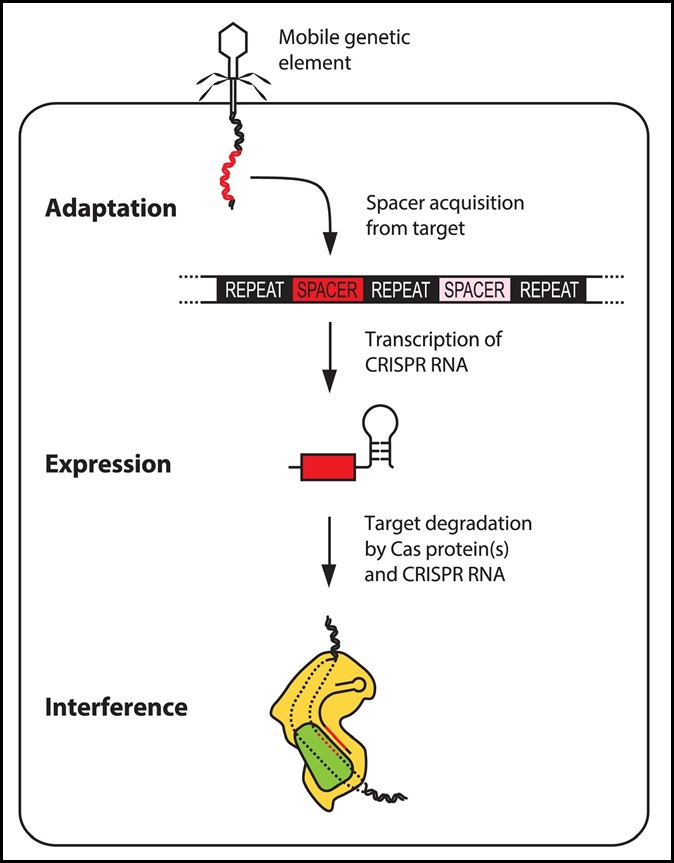
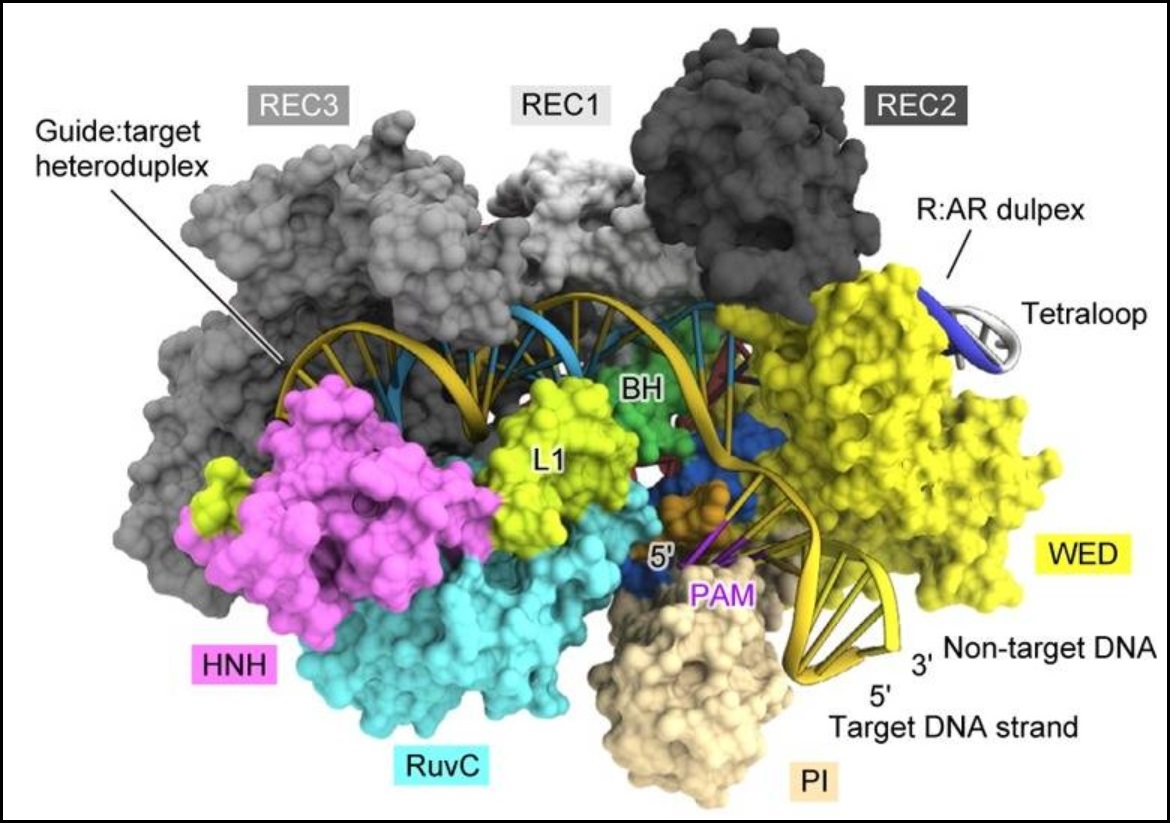
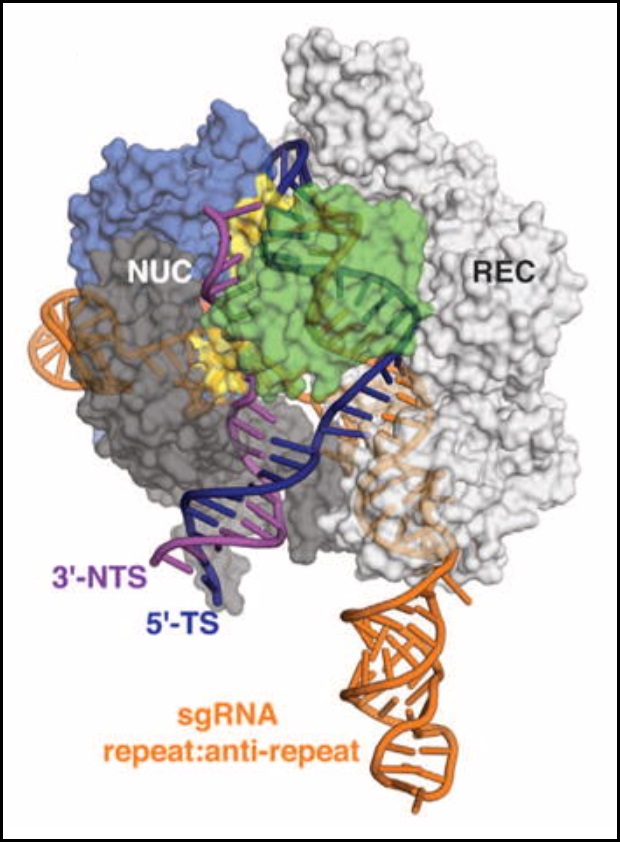
Cas9 is a bacterial and archaeal endonuclease that functions in CRISPR adaptive immunity. As part of CRISPR type-II systems, Cas9 recognizes protospacer-adjacent motif (PAM) sequences (usually 5'-NGG-3') in viral DNA, and then helps to integrate the viral sequence into a CRISPR array in the microbial genome. The CRISPR arrays are transcribed and processed, also involving Cas9, to create CRISPR RNAs (crRNAs). These crRNAs combine with tracrRNAs to form an RNA duplex. Cas9 binds the crRNA:tracrRNA duplex, and it uses the crRNA as a guide to bind and cleave both strands of a complementary viral sequence. Since 2012, Cas9 has become famous for its applications in highly-specific gene editing, in combination with sgRNAs that simulate crRNA:tracrRNA duplexes..



In this tutorial, we examine:.
> The Cas9 of Francisella novicida (FnCas9), a large and well-crystallized structure, plus bound DNA and sgRNA
> The Cas9 of Streptococcus pyogenes (SpCas9), commonly used in gene editing, plus bound DNA and sgRNA
> Comparisons between FnCas9 and SpCas9 to visualize structural homology
> CONSURF homology modeling based on SpCas9 to reveal regions of high and low conservation
| Francisella novicida Cas9 | Page 1 |
| Streptococcus pyogenes Cas9 | Page 2 |
| FnCas9 and SpCas9 comparison | Page 3 |
| SpCas9 homology model | Page 4 |