
Publications
Google Scholar Profile
 Orcid.org/0000-0002-2488-0239
Orcid.org/0000-0002-2488-0239
The PDF copies provided on this page are for preview only. Downloading is not permitted.
Publications Affiliated with SMU:
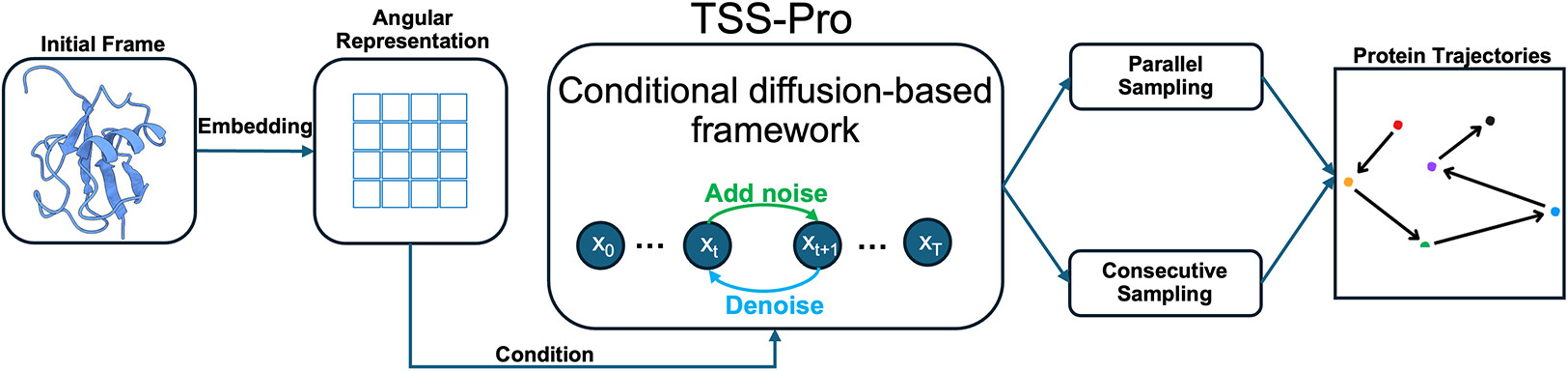
86. Xiong, C.; Kandhan, P.; Chen, D.; Ma, Z.; Smith, E. D.; Tao, P.*; Efficient Sampling of Short Protein Trajectories with Conditional Diffusion Models. 2026, J. Chem. Theory Comput. 22, 1, 78–94. DOI: 10.1021/acs.jctc.5c01579
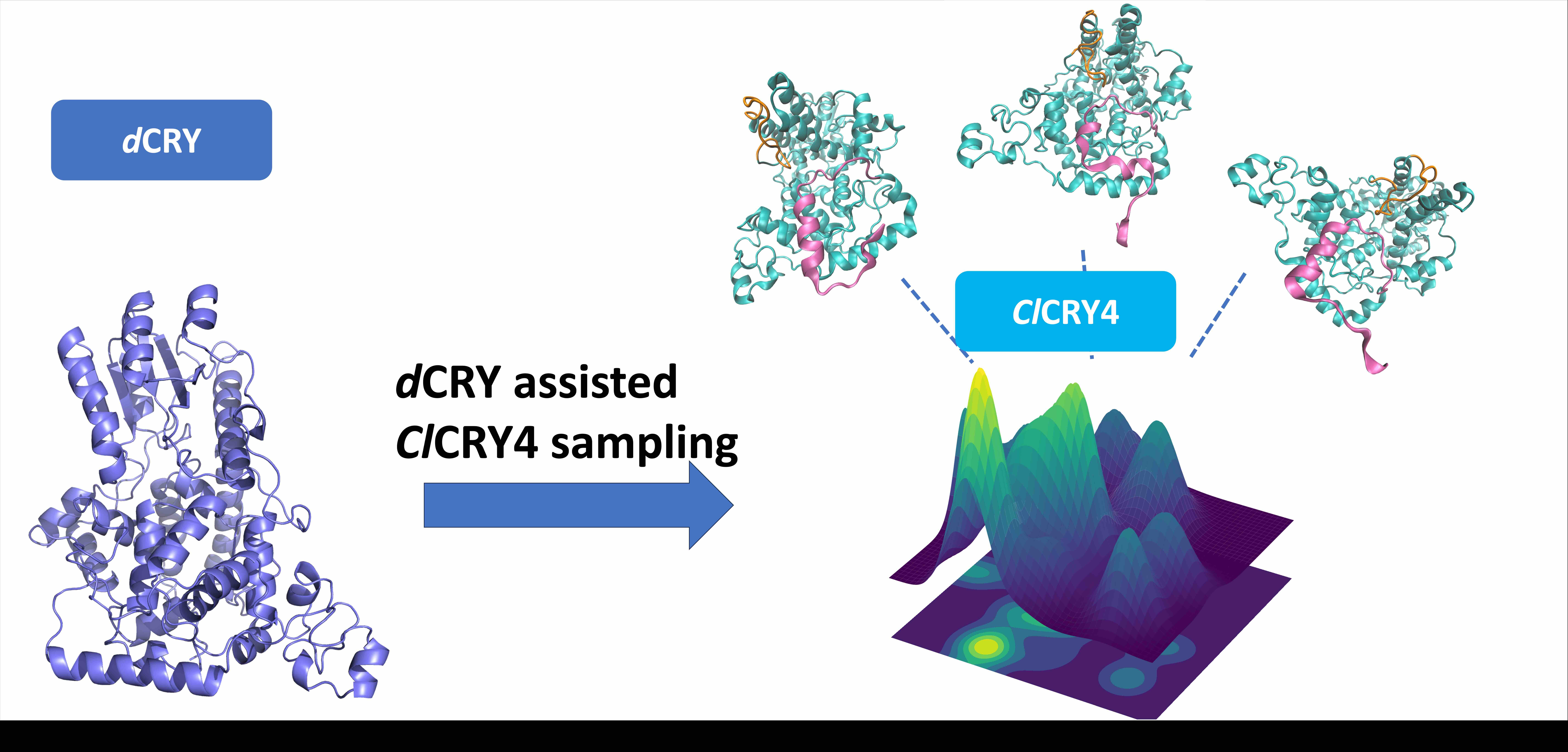
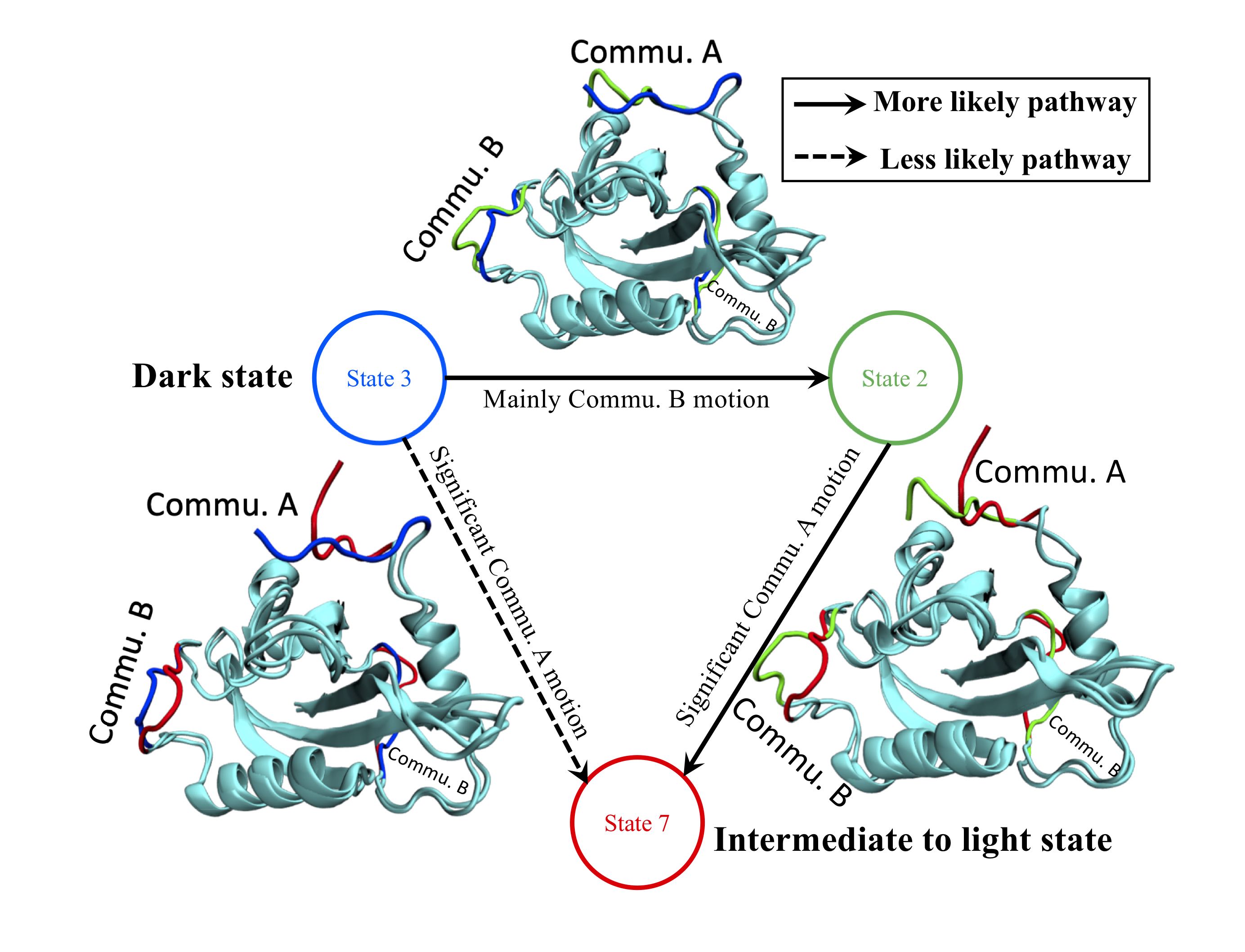
85. Xiong, C.; Kandhan, P.; Zoltowski, B. D.; Tao, P.*; Structural Plasticity and Functional Dynamics of Pigeon Cryptochrome 4 as Avian Magnetoreceptor. 2025, J. Mol. Biol. 437, 169233 DOI: 10.1016/j.jmb.2025.169233
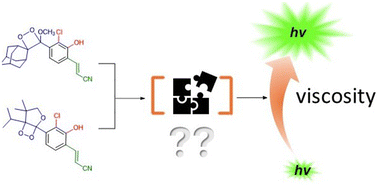
84. Cabello, M.; Kandhan, P.; Tao, P.; Lippert, A. R.*; Viscosity effects on the chemiluminescence emission of 1,2-dioxetanes in water. 2025, Org. Biomol. Chem., 23, 5380-5389 DOI: 10.1039/d5ob00254k
83. Amaro, R. E.; et al; The need to implement FAIR principles in biomolecular simulations. 2025, Nature Methods, 22, 641-645 DOI: 10.1038/s41592-025-02635-0
82. Xiao, S.; Alshahrani, M.; Hu, G.; Tao, P.; Verkhivker, G. M.;* Exploring binding and allosteric energy landscapes for the KRAS interactions with effector proteins using Markov state modeling of conformational ensembles and allosteric network modeling. 2025, Protein Science, 34:e70228. DOI: 10.1002/pro.70228
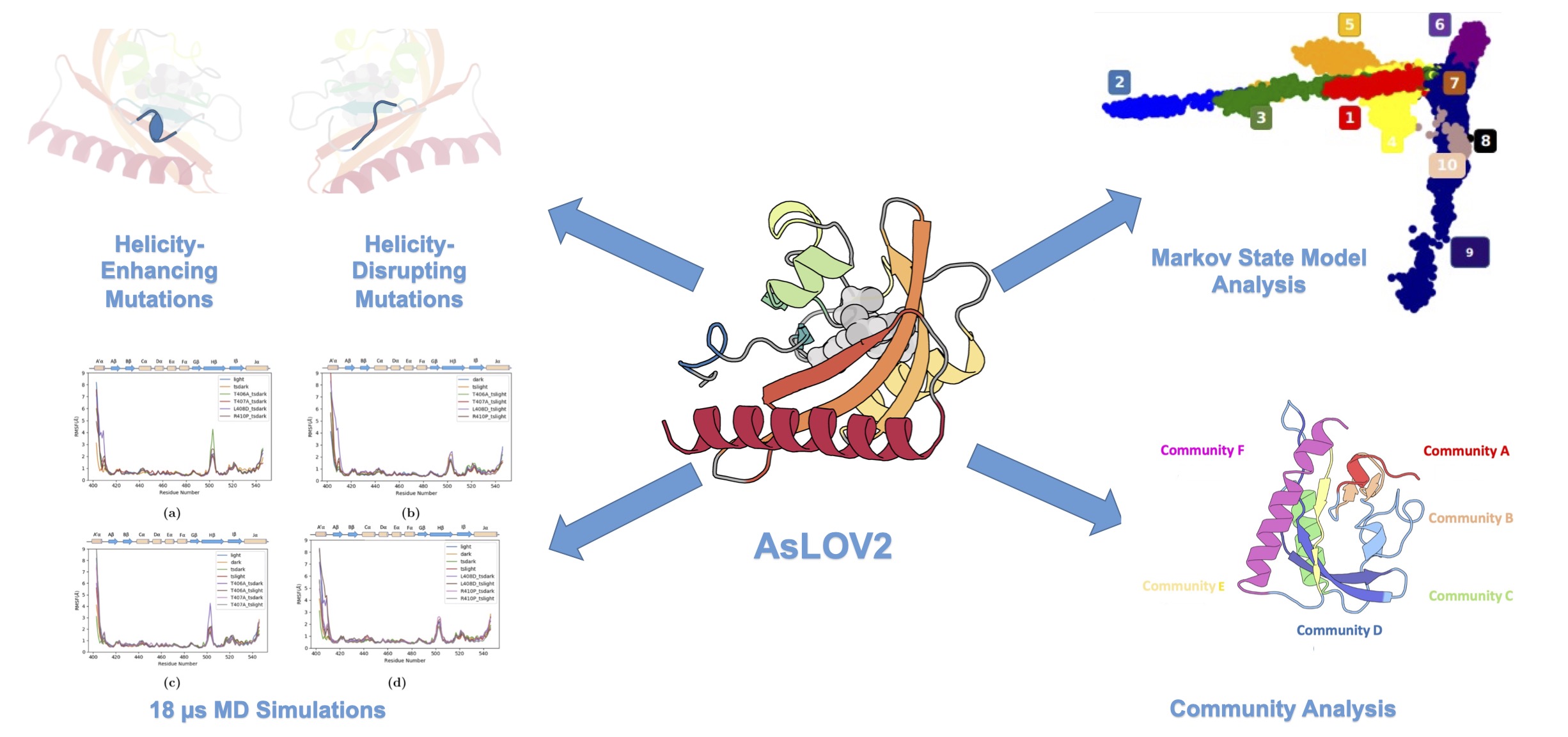
81. Xiao, S.; Ibrahim, M. T.; Verkhivker, G. M.; Zoltowski, B. D; Tao, P.* β-Sheets mediate the conformational change and allosteric signal transmission between the AsLOV2 termini. 2024, J. Comput. Chem. 45, 1493 DOI: 10.1002/jcc.27344

80. Raisinghani, N.; Alshahrani, M.; Gupta, G.;Tian, H.; Xiao, S.; Tao, P.*; Verkhivker, G. M.*; Integration of a Randomized Sequence Scanning Approach in AlphaFold2 and Local Frustration Profiling of Conformational States Enable Interpretable Atomistic Characterization of Conformational Ensembles and Detection of Hidden Allosteric States in the ABL1 Protein Kinase. 2024, J. Chem. Theory Comput. 20, 5317. DOI: 10.1021/acs.jctc.4c00222

79. Raisinghani, N.; Alshahrani, M.; Gupta, G.; Tian, H.; Xiao, S.; Tao, P.; Verkhivker, G. M.*; Probing Functional Allosteric States and Conformational Ensembles of the Allosteric Protein Kinase States and Mutants: Atomistic Modeling and Comparative Analysis of AlphaFold2, OmegaFold, and AlphaFlow Approaches and Adaptations. 2024, J. Phys. Chem. B, 2024, 128, 11088. DOI: 10.1021/acs.jpcb.4c04985

78. Raisinghani, N.; Alshahrani, M.; Gupta, G.; Tian, H.; Xiao, S.; Tao, P.; Verkhivker, G. M.*; AlphaFold2-Enabled Atomistic Modeling of Structure, Conformational Ensembles, and Binding Energetics of the SARS-CoV-2 Omicron BA.2.86 Spike Protein with ACE2 Host Receptor and Antibodies: Compensatory Functional Effects of Binding Hotspots in Modulating Mechanisms of Receptor Binding and Immune Escape. 2024, J. Chem. Inf. Model. 2024, 64, 1657. DOI: 10.1021/acs.jcim.3c01857
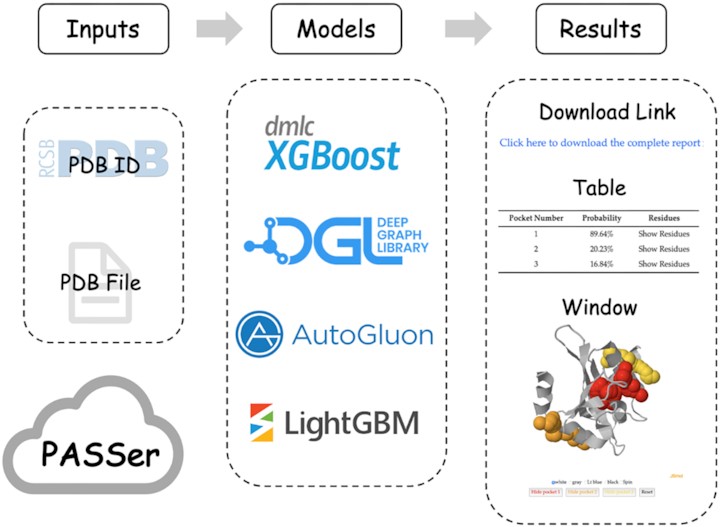
77. Tian, H.; Xiao, S.; Jiang. X; Tao, P.* PASSer: fast and accurate prediction of protein allosteric sites 2023, Nucleic Acids Research. 51 (W1), W427–W431 DOI: 10.1093/nar/gkad303
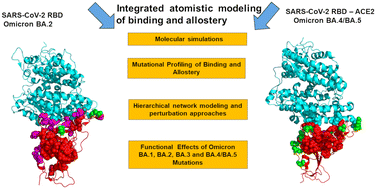
76. Verkhivker, G. M.*; Alshahrani, M.; Gupta, G.; Xiao, S.; and Tao, P. Probing conformational landscapes of binding and allostery in the SARS-CoV-2 omicron variant complexes using microsecond atomistic simulations and perturbation-based profiling approaches: hidden role of omicron mutations as modulators of allosteric signaling and epistatic relationships. 2023, Phys. Chem. Chem. Phys. 25, 21245. DOI: 10.1039/d3cp02042h
75. Tian, H.; Xiao, S.; Jiang. X; Tao, P.* PASSerRank: Prediction of allosteric sites with learning to rank 2023, J. Comput. Chem. 44, 2223. DOI: 10.1002/jcc.27193

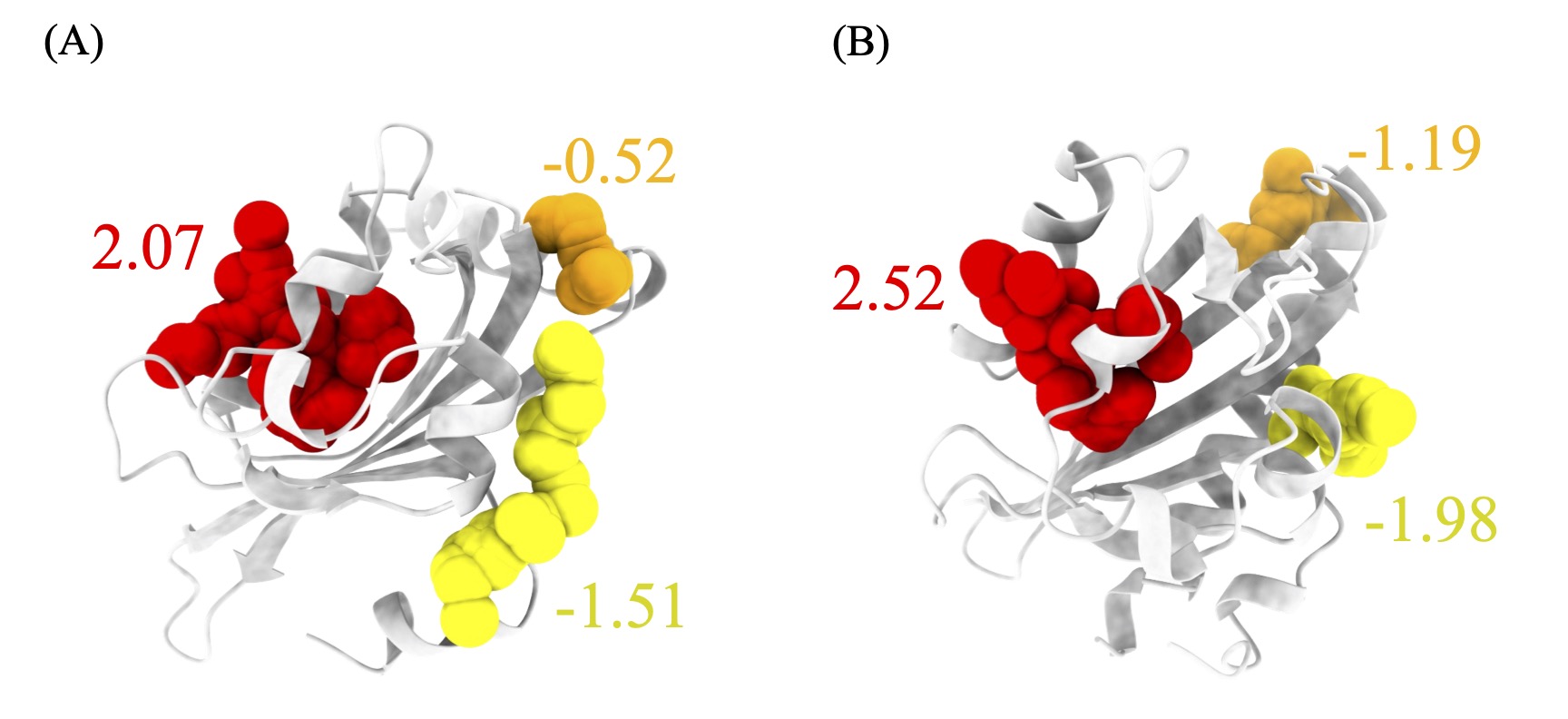
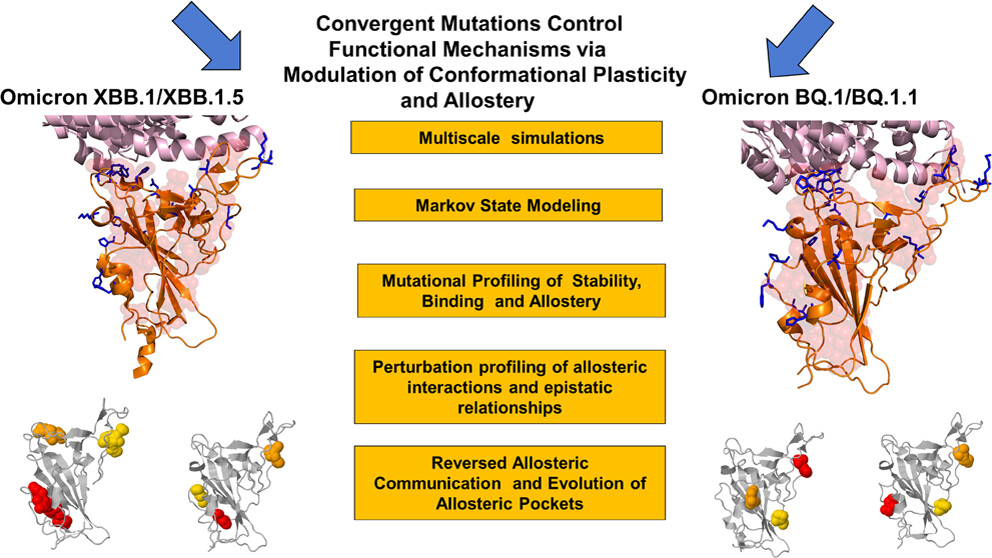
74. Xiao, S.; Alshahrani, M.; Gupta, G.; and Tao, P.*; Verkhivker, G. M.* Markov State Models and Perturbation-Based Approaches Reveal Distinct Dynamic Signatures and Hidden Allosteric Pockets in the Emerging SARS-Cov-2 Spike Omicron Variant Complexes with the Host Receptor: The Interplay of Dynamics and Convergent Evolution Modulates Allostery and Functional Mechanisms 2023, J. Chem. Inf. Model. 63, 5272. DOI: 10.1021/acs.jcim.3c00778
73. Xiao, S.; Song, Z.; Tian, H.; Tao, P.* Assessments of Variational Autoencoder in Protein Conformation Exploration 2023, Journal of Computational Biophysics and Chemistry. 22 (4), 489-501. DOI: 10.1142/S2737416523500217
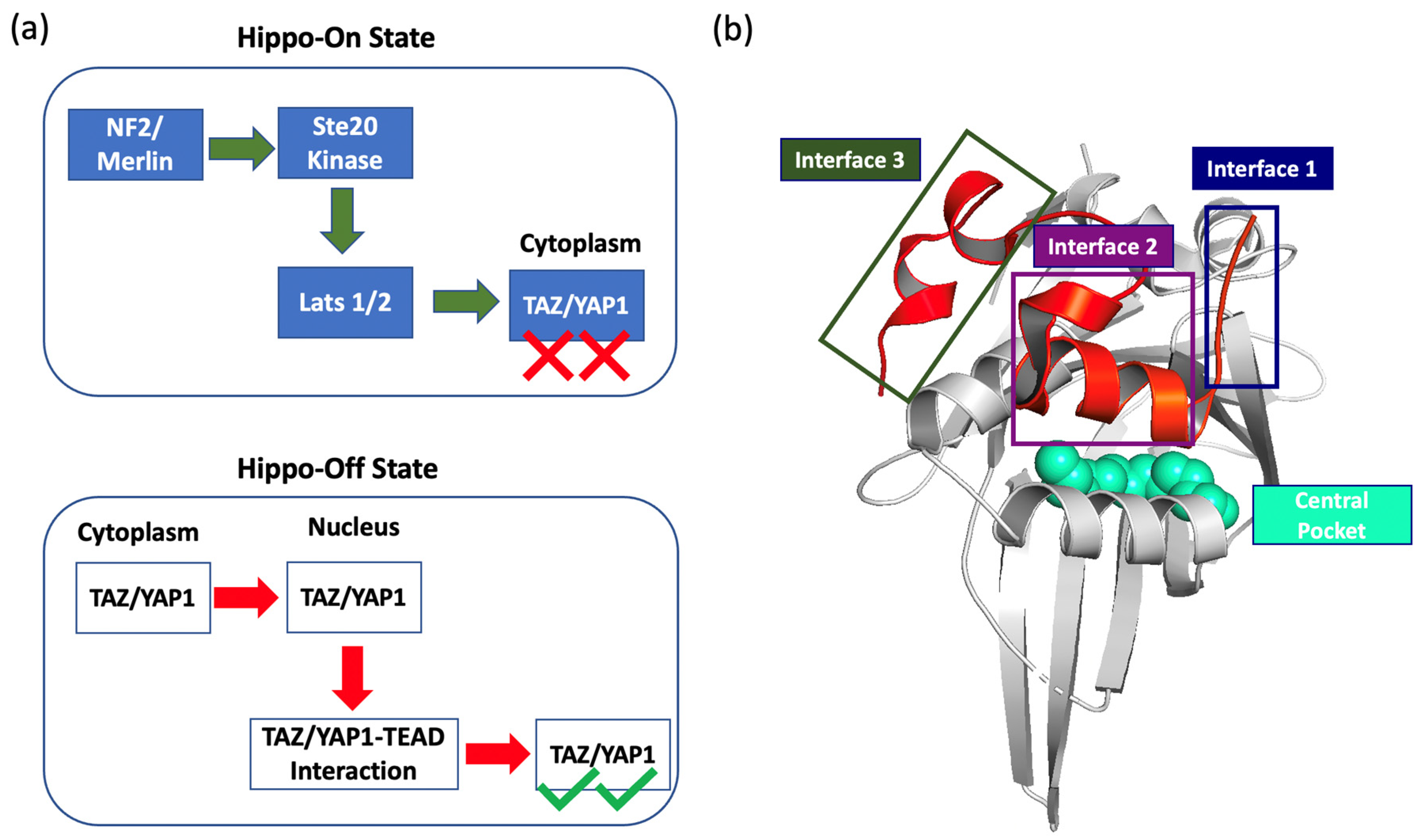
72. Ibrahim, M. T.; Verkhivker, G. M.; Misra, J.; Tao, P.* Novel Allosteric Effectors Targeting Human Transcription Factor TEAD 2023, Int. J. Mol. Sci. 24 (10), 9009 DOI: 10.3390/ijms24109009
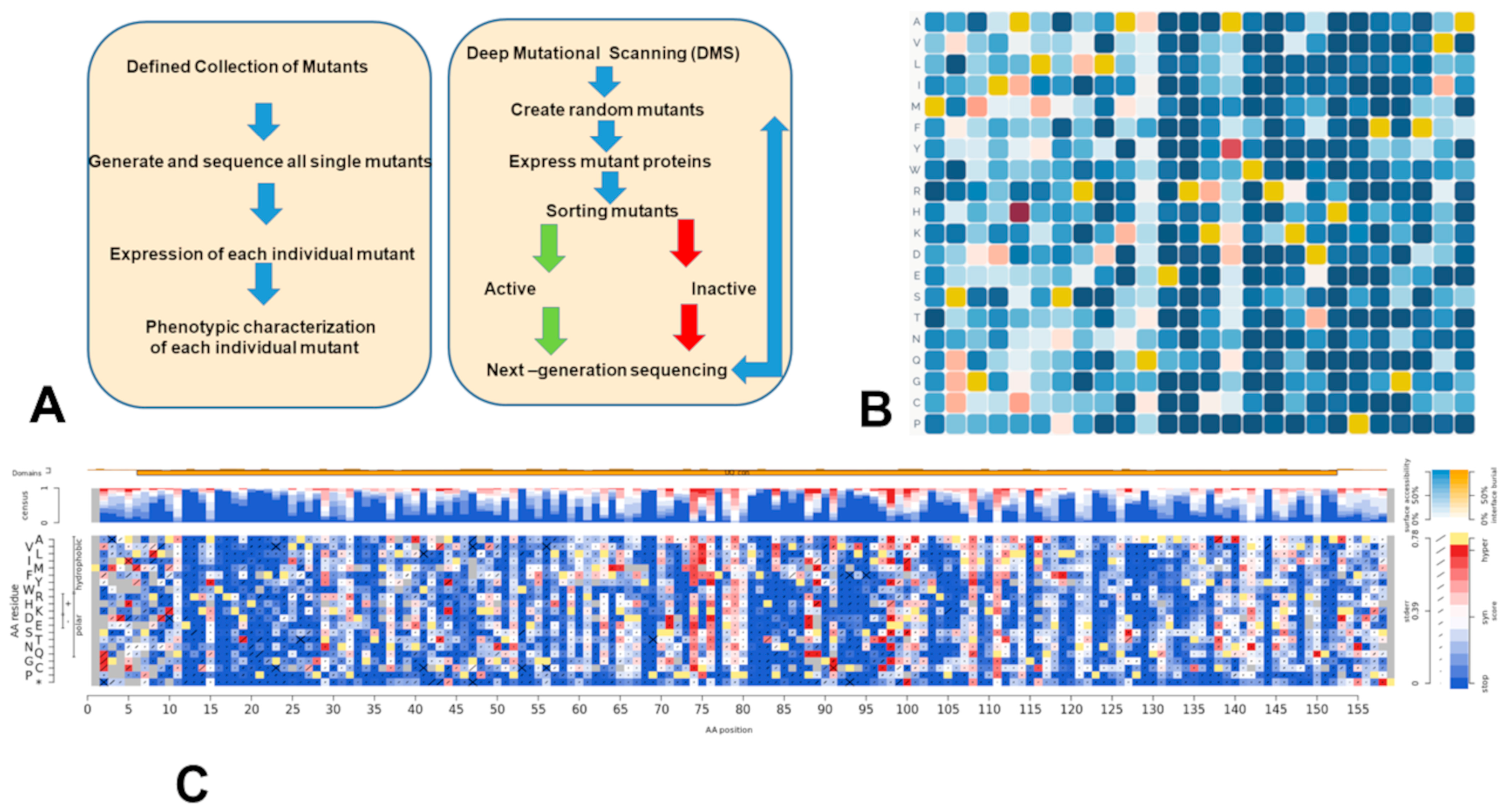
71. Verkhivker, G. M.*; Alshahrani, M.; Gupta, G.; Xiao, S.; and Tao, P. From Deep Mutational Mapping of Allosteric Protein Landscapes to Deep Learning of Allostery and Hidden Allosteric Sites: Zooming in on “Allosteric Intersection” of Biochemical and Big Data Approaches. 2023, Int. J. Mol. Sci 24 (9), 7747. DOI: 10.3390/ijms24097747
70. Agajanian, S.; Alshahrani, M.; Bai, F.; Tao, P.; Verkhivker, G. M.* Exploring and Learning the Universe of Protein Allostery Using Artificial Intelligence Augmented Biophysical and Computational Approaches. 2023, J. Chem. Inf. Model. 63, 1413. DOI: 10.1021/acs.jcim.2c01634
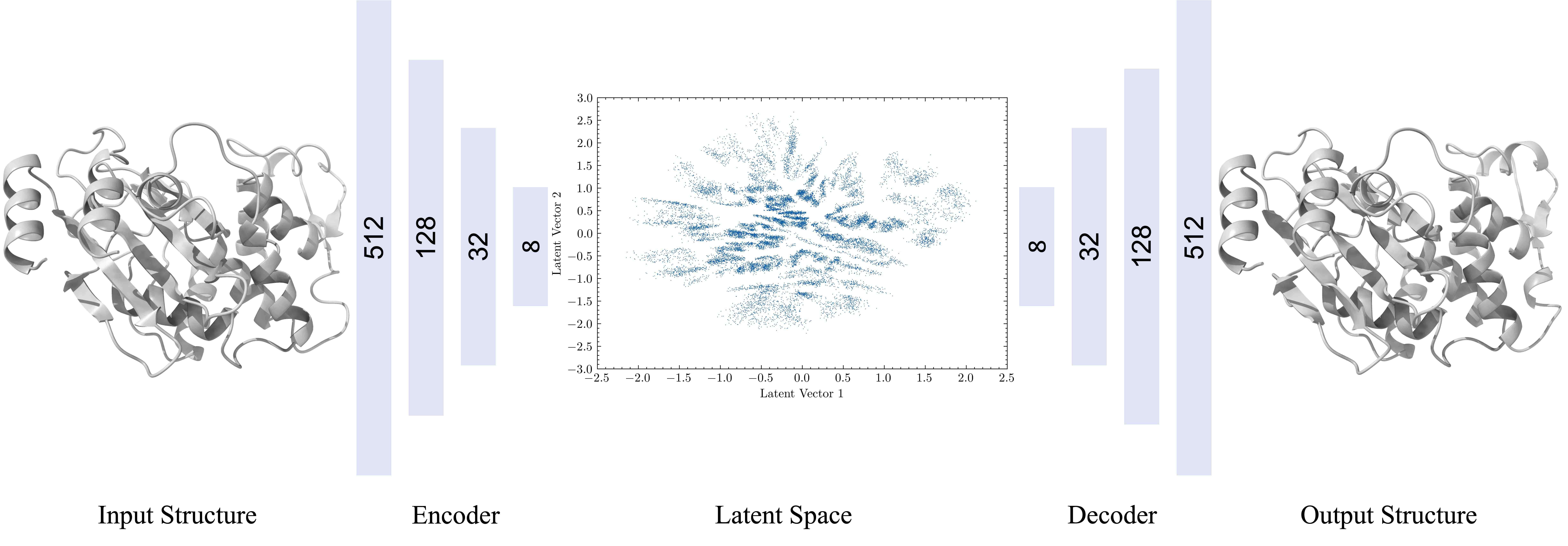
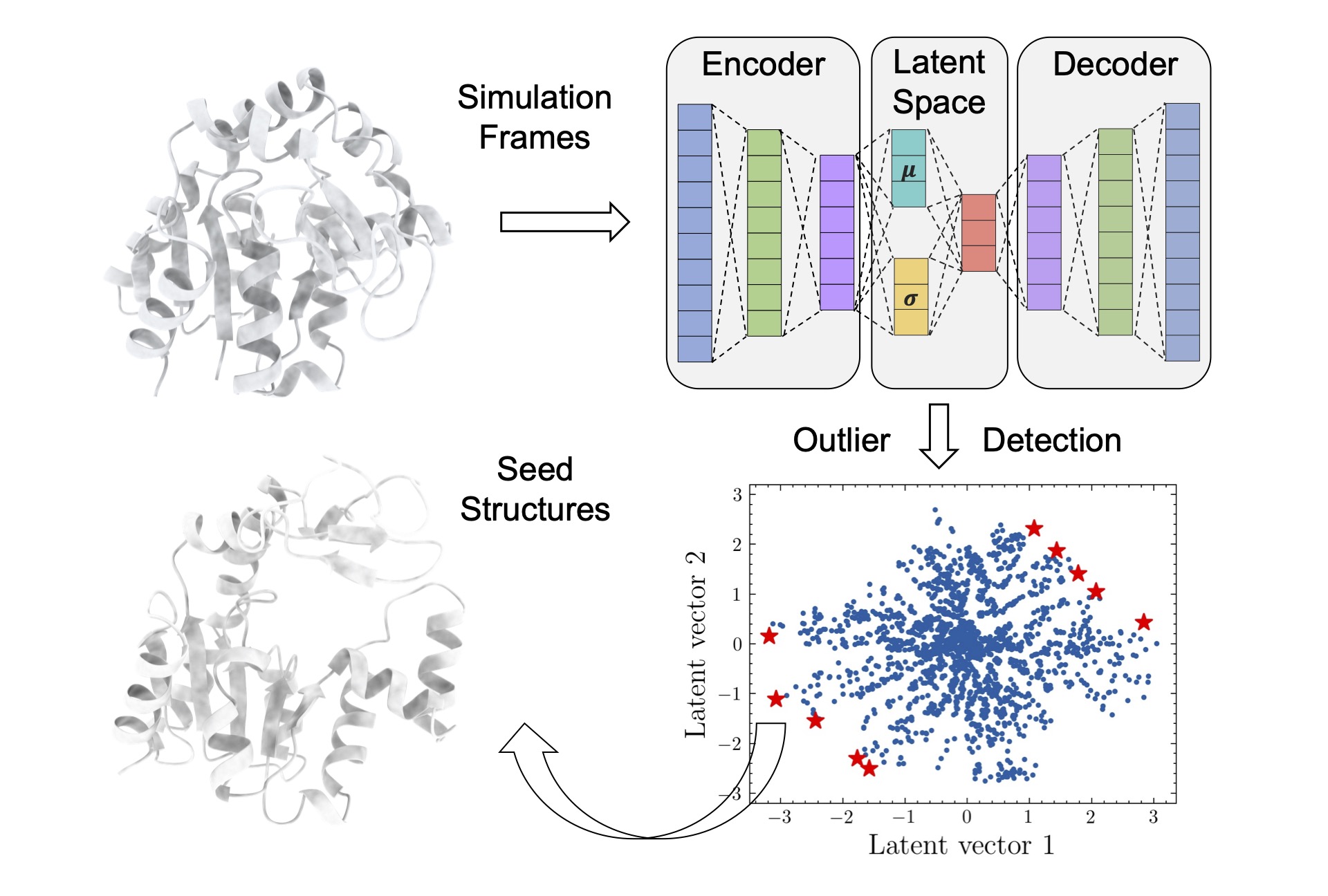
69. Tian, H.; Jiang. X; Xiao, S.; La Force, H.; Larson, E. C.; Tao, P.* LAST: Latent Space-Assisted Adaptive Sampling for Protein Trajectories. 2023, J. Chem. Inf. Model. 63, 67. DOI: 10.1021/acs.jcim.2c01213
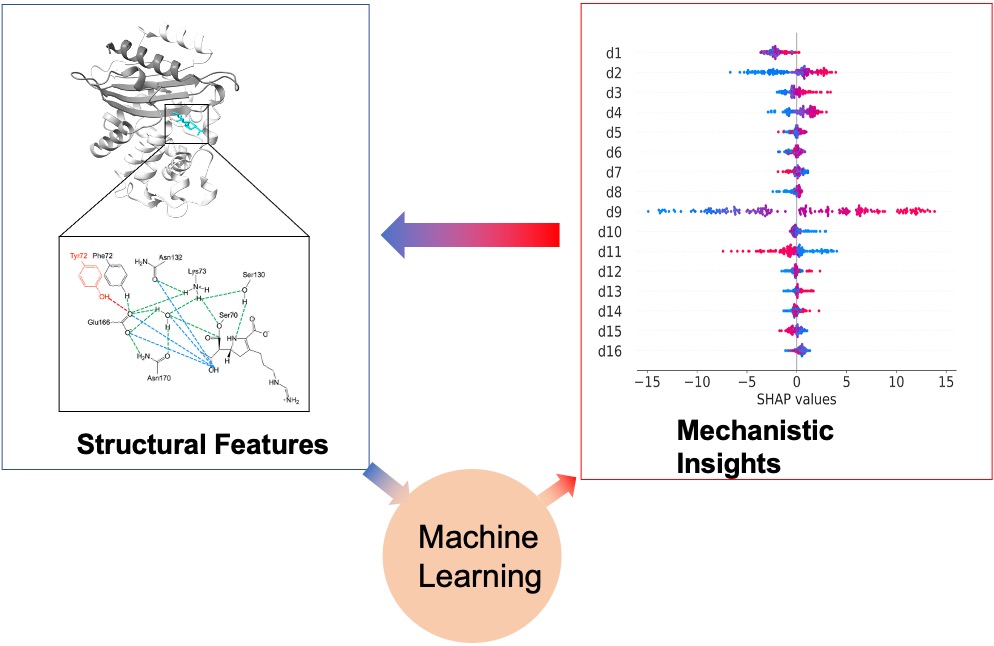
68. Yin, C.; Song, Z.; Tian, H.; Palzkill, T.; Tao, P.* Unveiling the structural features that regulate carbapenem deacylation in KPC-2 through QM/MM and interpretable machine learning. 2023, Phys. Chem. Chem. Phys. 25, 1349. DOI: 10.1039/d2cp03724f
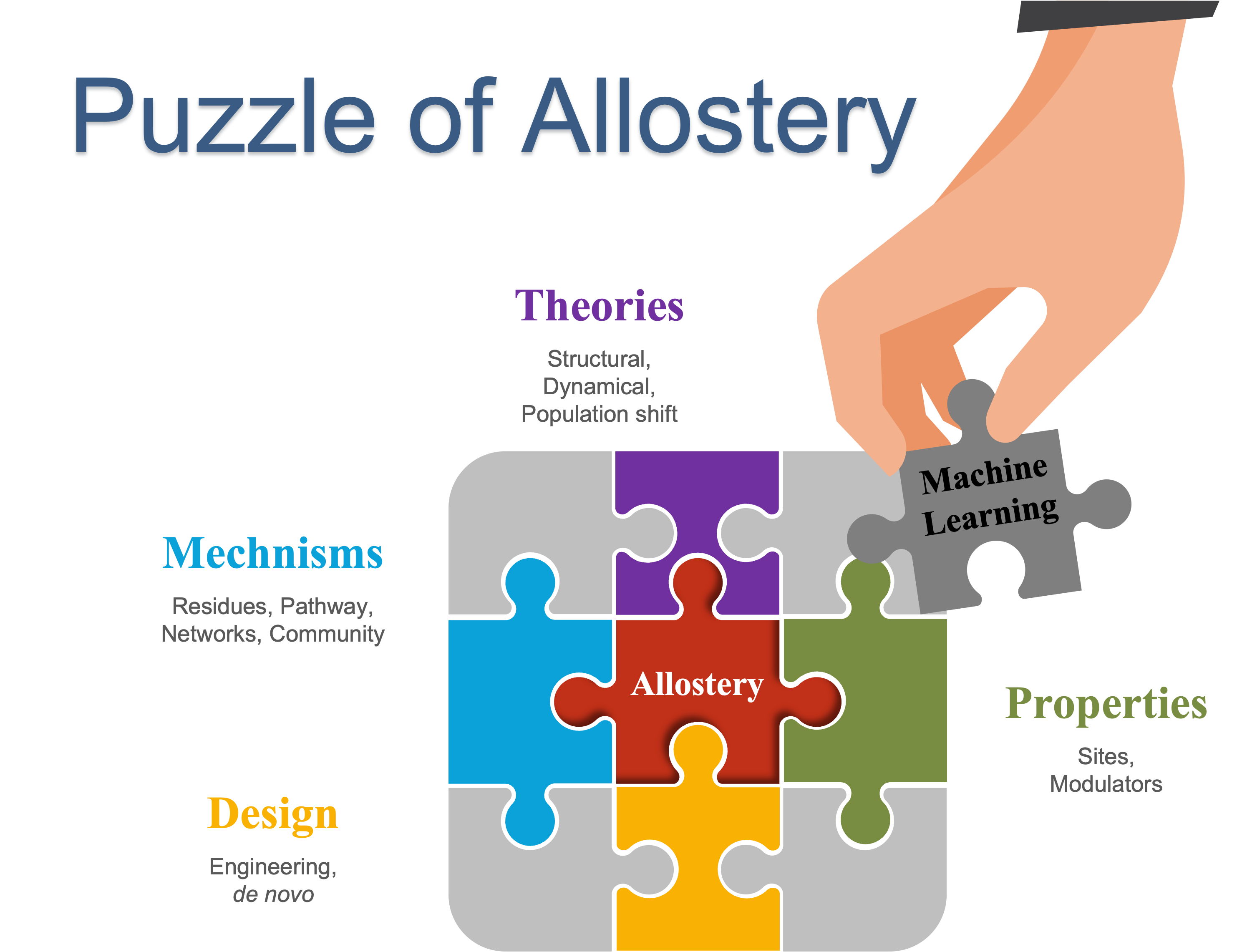
67. Xiao, S.; Verkhivker, G.; Tao, P.* Machine learning and protein allostery. 2022, Trends in Biochemical Sciences. 48, 375. DOI: 10.1016/j.tibs.2022.12.001
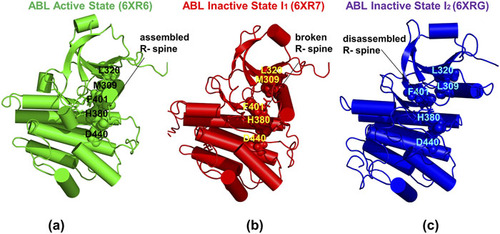
66. Krishnan, K.; Tian, H.; Tao, P.; Verkhivker, G. Probing conformational landscapes and mechanisms of allosteric communication in the functional states of the ABL kinase domain using multiscale simulations and network-based mutational profiling of allosteric residue potentials. 2022, J. Chem. Phys. 157, 245101. DOI: 10.1063/5.0133826
65. Tian, H.; Ketkar, R.; Tao, P.* ADMETboost: a web server for accurate ADMET prediction. Journal of Molecular Modeling. 2022, 28:408. DOI: 10.1007/s00894-022-05373-8
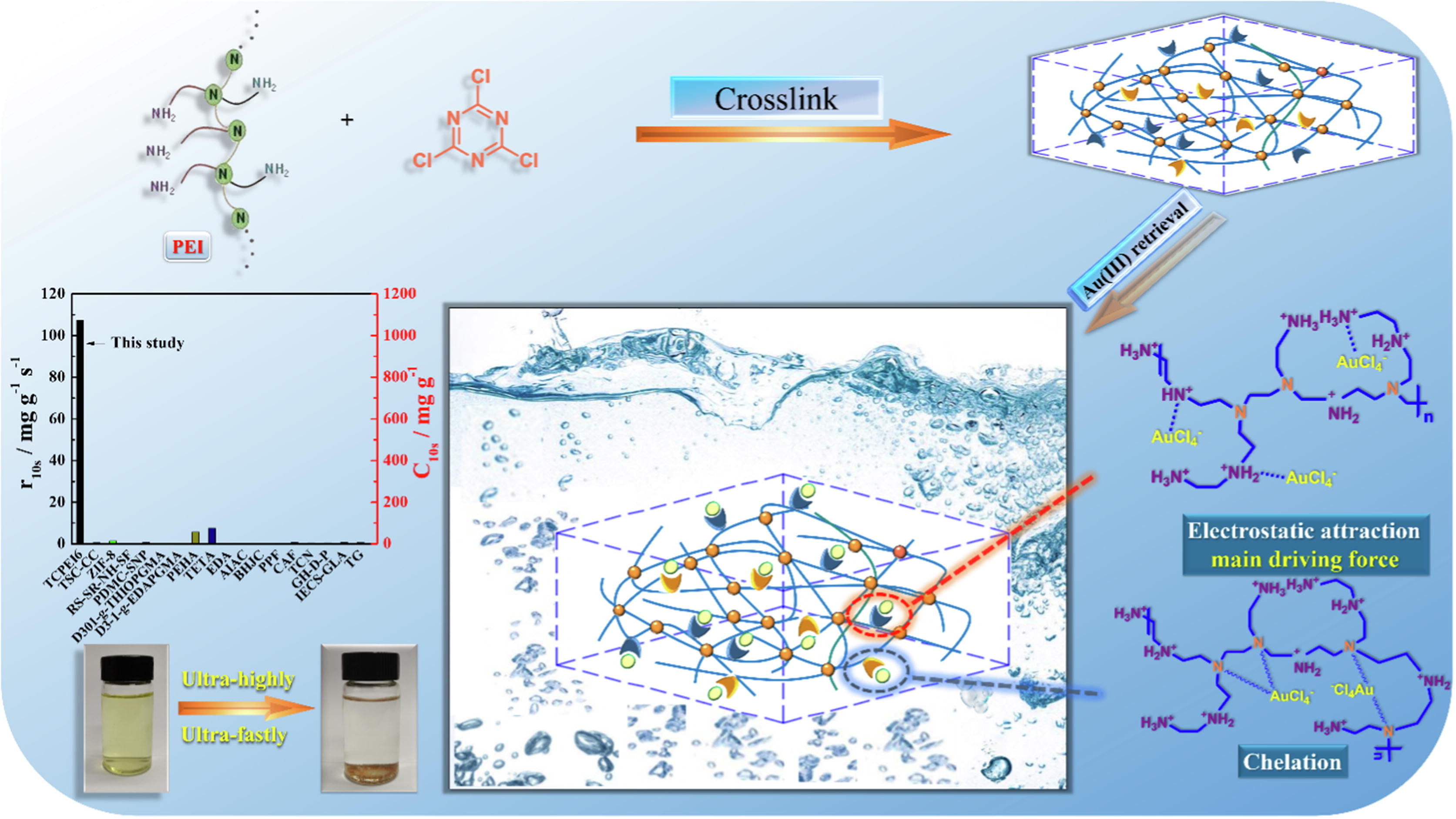
64. Hu, B.; Yang, M.; Huang, H.; Song, Z.; Tao, P. Wu, Y.; Tang, K.; Chen, X.; Yang, C. Triazine-crosslinked polyethyleneimine for efficient adsorption and recovery of gold from wastewater. 2022, Journal of Molecular Liquids. 367,120586. DOI: 10.1016/j.molliq.2022.120586
63. Song, Z.; Trozzi, F.; Tian, H.; Chao, Y.; Tao, P.* Mechanistic Insights into Enzyme Catalysis from Explaining Machine-Learned Quantum Mechanical and Molecular Mechanical Minimum Energy Pathways. 2022, ACS Phys. Chem Au, 2, 316. DOI: 10.1021/acsphyschemau.2c00005

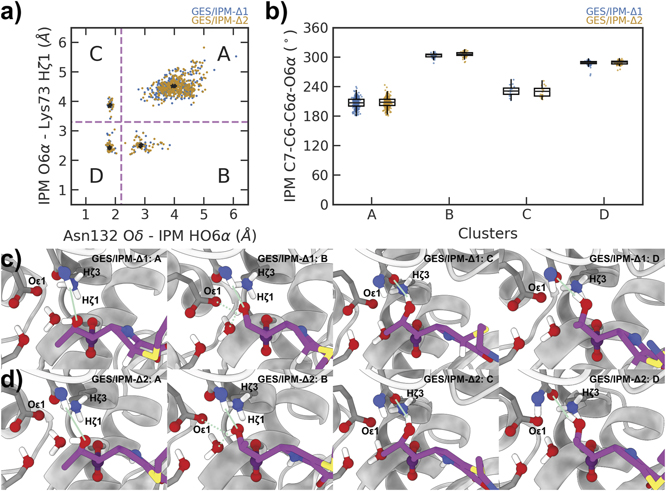
62. Song, Z.; Tao, P.* Graph-learning guided mechanistic insights into imipenem hydrolysis in GES carbapenemases. 2022, Electron. Struct. 4, 034001. DOI: 10.1088/2516-1075/ac7993
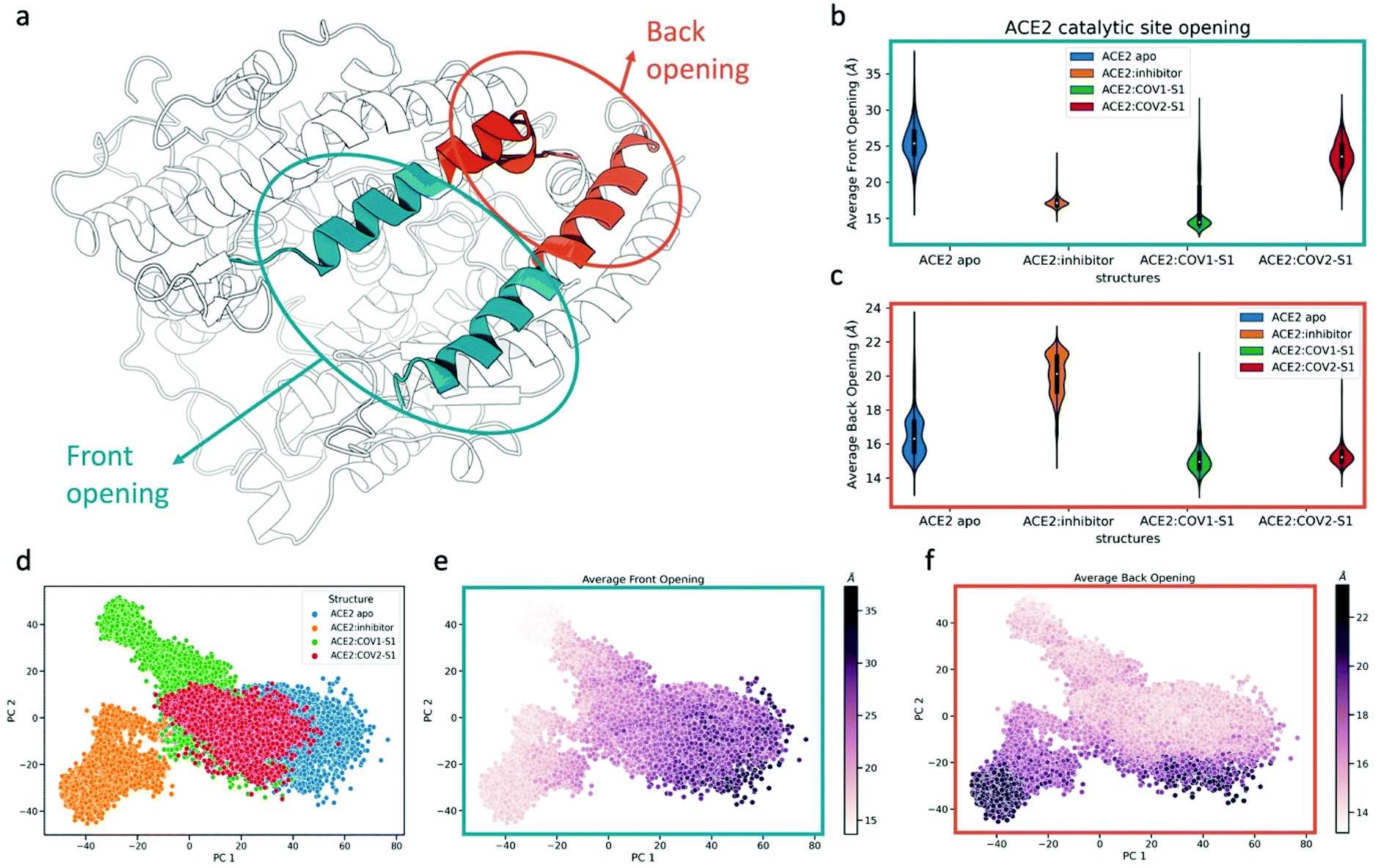
61. Trozzi, F.; Karki, N.; Song, Z.; Verma, N.; Kraka, E.; Zoltowski, B. D.; Tao, P.* Allosteric control of ACE2 peptidase domain dynamics. 2022, Org. Biomol. Chem. 20, 3605. DOI: 10.1039/d2ob00606e
60. Ibrahim, M. T.; Lee, J.; Tao, P* Homology modeling of Forkhead box protein C2: identification of potential inhibitors using ligand and structure‐based virtual screening. 2022, Molecular Diversity, 27, 1661. DOI: 10.1007/s11030-022-10519-0
59. Ibrahim, M. T.; Tao, P* Computational investigation of peptidomimetics as potential inhibitors of SARS-CoV-2 spike protein. 2022, 41, 7144. Journal of Biomolecular Structure and Dynamics. DOI: 10.1080/07391102.2022.2116601
58. Xiao S.; Tian H.; Tao, P* PASSer2.0: Accurate Prediction of Protein Allosteric Sites Through Automated Machine Learning. 2022, Frontiers in Molecular Biosciences. 9:879251. DOI: 10.3389/fmolb.2022.879251
57. Bai, F.; Puk, K. M.; Liu, J.; Zhou, H.; Tao, P.; Zhou, W.; Wang, S.* Sparse group selection and analysis of function-related residue for protein-state recognition. 2022, J Comput Chem. 43: 1342–1354. DOI: 10.1002/jcc.26937
56. Ibrahim, M. T.; Trozzi, F.; Tao, P* Dynamics of hydrogen bonds in the secondary structures of allosteric protein Avena Sativa phototropin 1. 2021, Computational and Structural Biotechnology Journal. 20, 50-64. DOI: 10.1016/j.csbj.2021.11.038
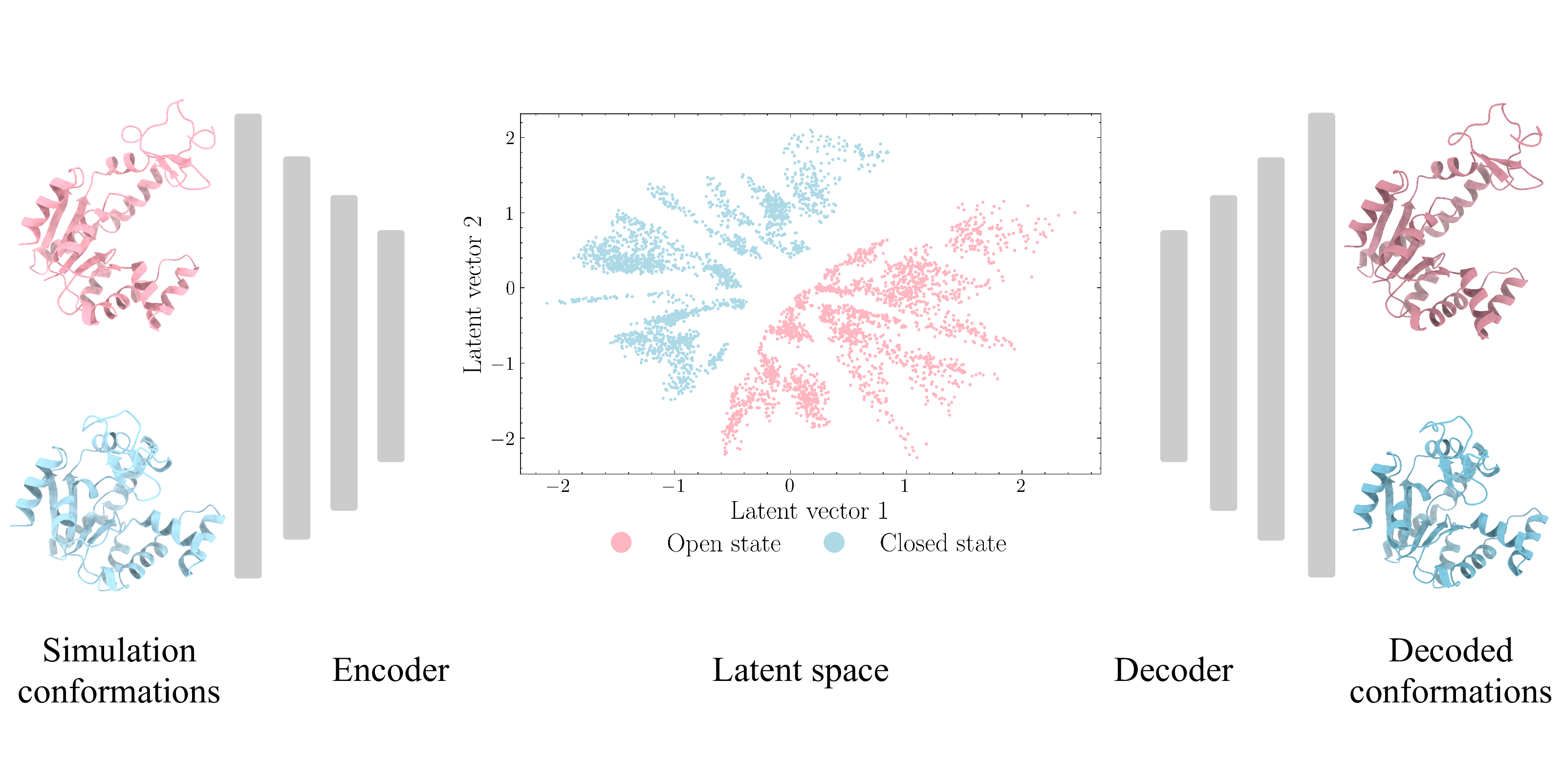
55. Tian H.; Jiang X.; Trozzi F.; Xiao S.; Larson E.; Tao P.* Explore protein conformational space with variational autoencoder. Front. Mol. Biosci. 8:781635. DOI: 10.3389/fmolb.2021.781635
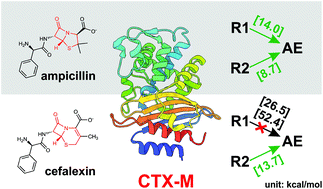
54. Song, Z.; Trozzi, F.; Palzkill, T.; Tao, P.* QM/MM modeling of class A β-lactamases reveals distinct acylation pathways for ampicillin and cefalexin, 2021, Org. Biomol. Chem., 19, 9182-9189. DOI: 10.1039/d1ob01593a
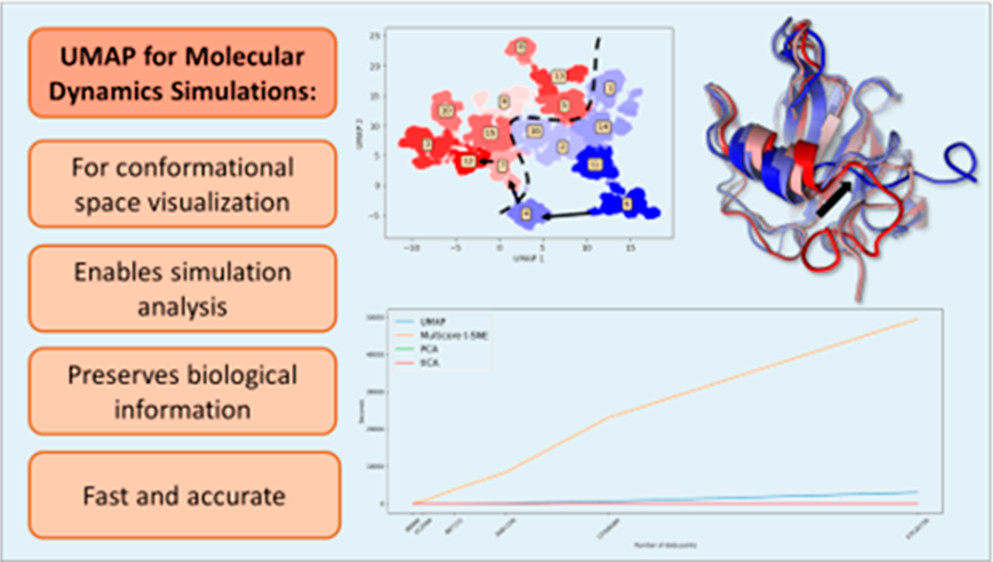
53. Trozzi, F.; Wang, X; Tao, P.* UMAP as a Dimensionality Reduction Tool for Molecular Dynamics Simulations of Biomacromolecules: A Comparison Study. 2021, J. Phys. Chem. B 125, 5022−5034. DOI: 10.1021/acs.jpcb.1c02081
52. Trozzi F.; Wang F.; Verkhivker G.; Zoltowski BD.; Tao P.* Dimeric allostery mechanism of the plant circadian clock photoreceptor ZEITLUPE. PLoS computational biology. 2021 Jul 26;17(7):e1009168. DOI: 10.1371/journal.pcbi.1009168
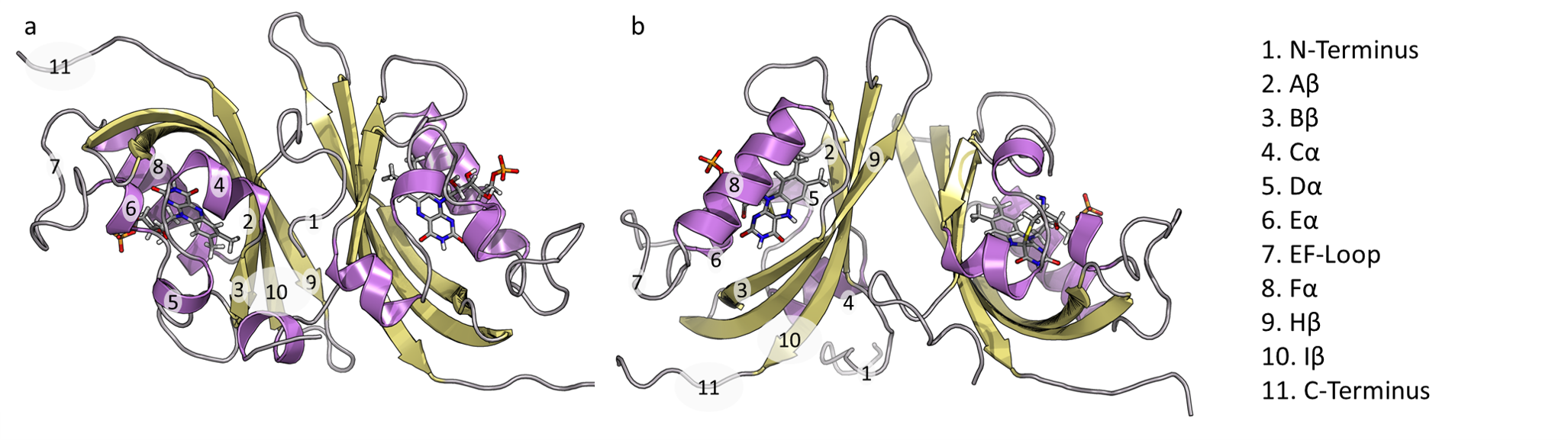
51. Tian, H.; Jiang, X.; Tao, P.* PASSer: Prediction of Allosteric Sites Server. 2021, Machine Learning: Science and Technology. 2, 035015 DOI: 10.1088/2632-2153.abe6d6 PASSer websever

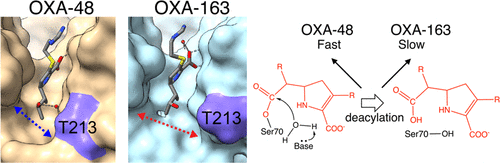
50. Stojanoski, V.; Hu, L.; Sankaran, B.; Wang, F.; Tao, P.; Prasad, B. V. V.; and Palzkill, T.* Mechanistic Basis of OXA-48-like β‐Lactamases’ Hydrolysis of Carbapenems. 2021, ACS Infect. Dis. 7, 445-460 DOI: 10.1021/acsinfecdis.0c00798
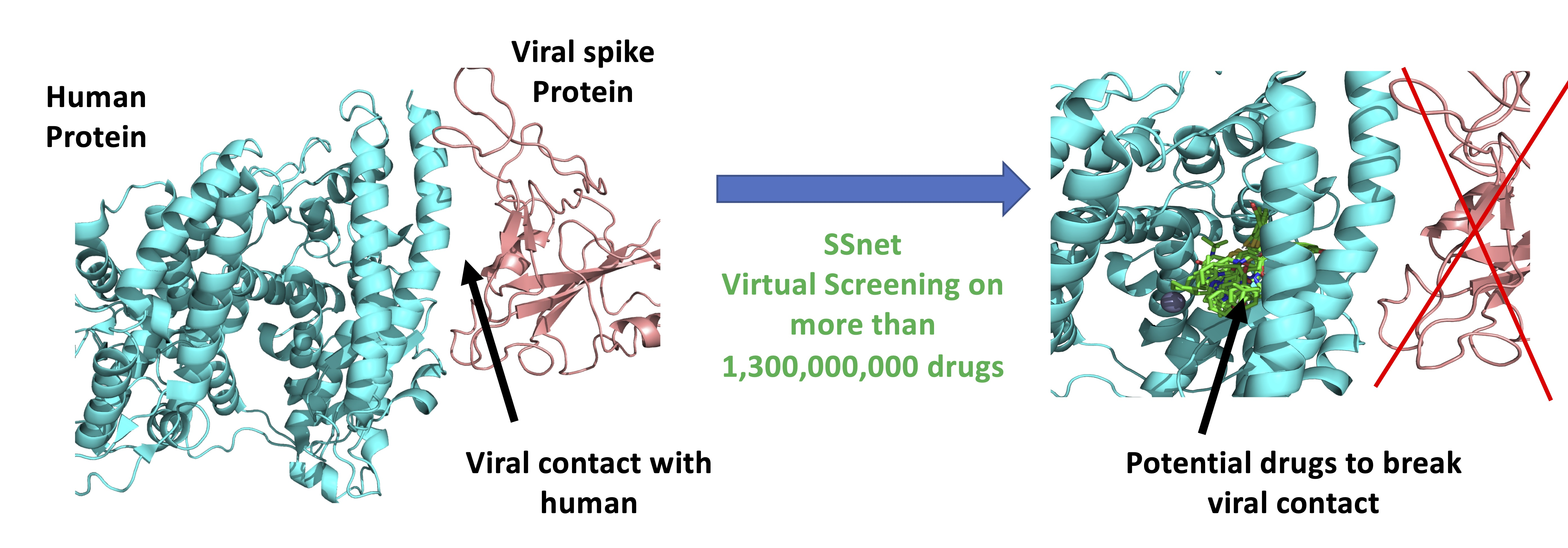
49. Karki, N.; Verma, N.; Trozzi, F.; Tao, P.; Kraka, E.; and Zoltowski, B. Predicting Potential SARS-COV-2 Drugs—In Depth Drug Database Screening Using Deep Neural Network Framework SSnet, Classical Virtual Screening and Docking. 2021, Int. J. Mol. Sci. 22, 1573 DOI: 10.3390/ijms22041573
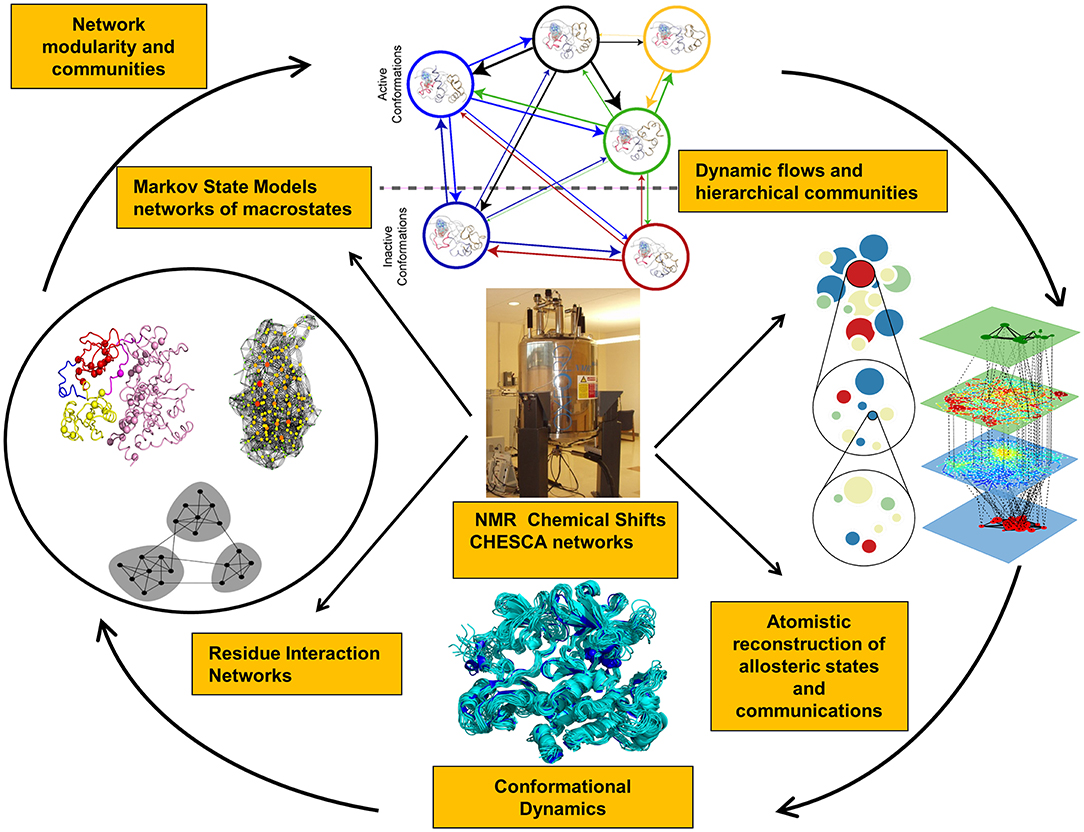
48. Verkhivker, G. M.*; Agajanian, S.; Hu, G.; and Tao, P. Allosteric Regulation at the Crossroads of New Technologies: Multiscale Modeling, Networks, and Machine Learning. 2020, Front. Mol. Biosci., 7, 136 DOI: 10.3389/fmolb.2020.00136
47. Song, Z.; Zhou, H.; Tian, H.; Wang, X.; Tao, P.* Unraveling the energetic significance of chemical events in enzyme catalysis via machine-learning based regression approach. 2020, Communications Chemistry 3, 134 DOI: 10.1038/s42004-020-00379-w
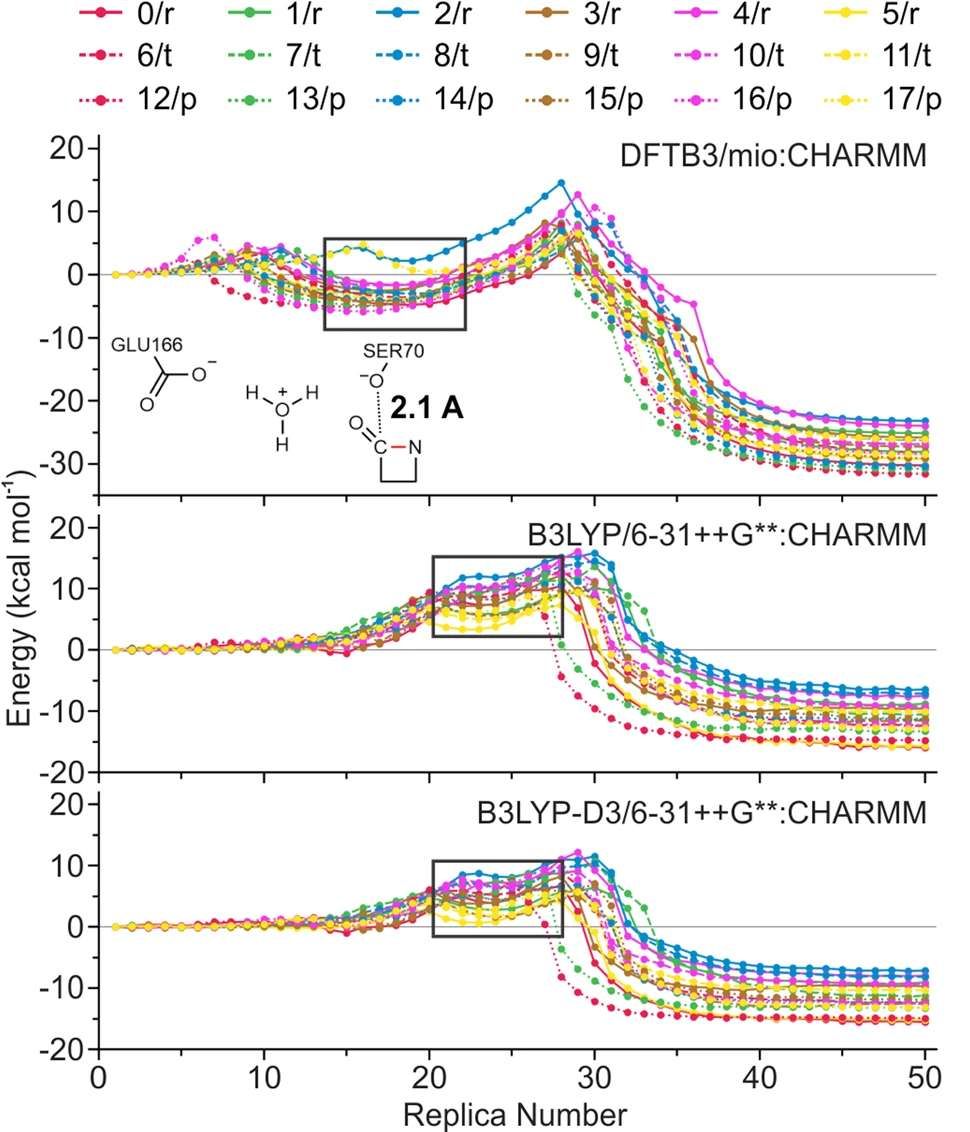
46. Tian, H.; Trozzi, F.; Zoltowski, B. D.; Tao, P.* Deciphering the Allosteric Process of the Phaeodactylum tricornutum Aureochrome 1a LOV Domain. 2020, J. Phys. Chem. B, 124, 41, 8960–8972 DOI: 10.1021/acs.jpcb.0c05842
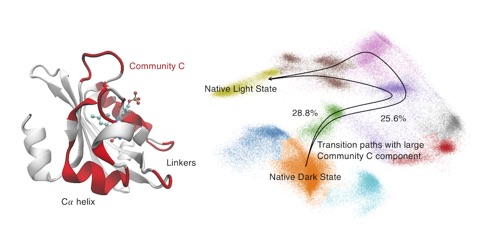
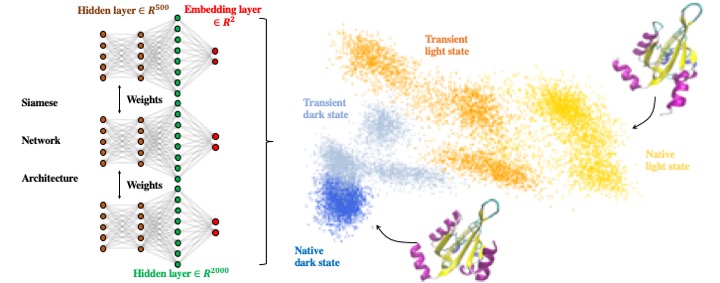
45. Tian, H.; Tao, P.* ivis Dimensionality Reduction Framework for Biomacromolecular Simulations. 2020, J. Chem. Inf. Model. DOI: 10.1021/acs.jcim.0c00485
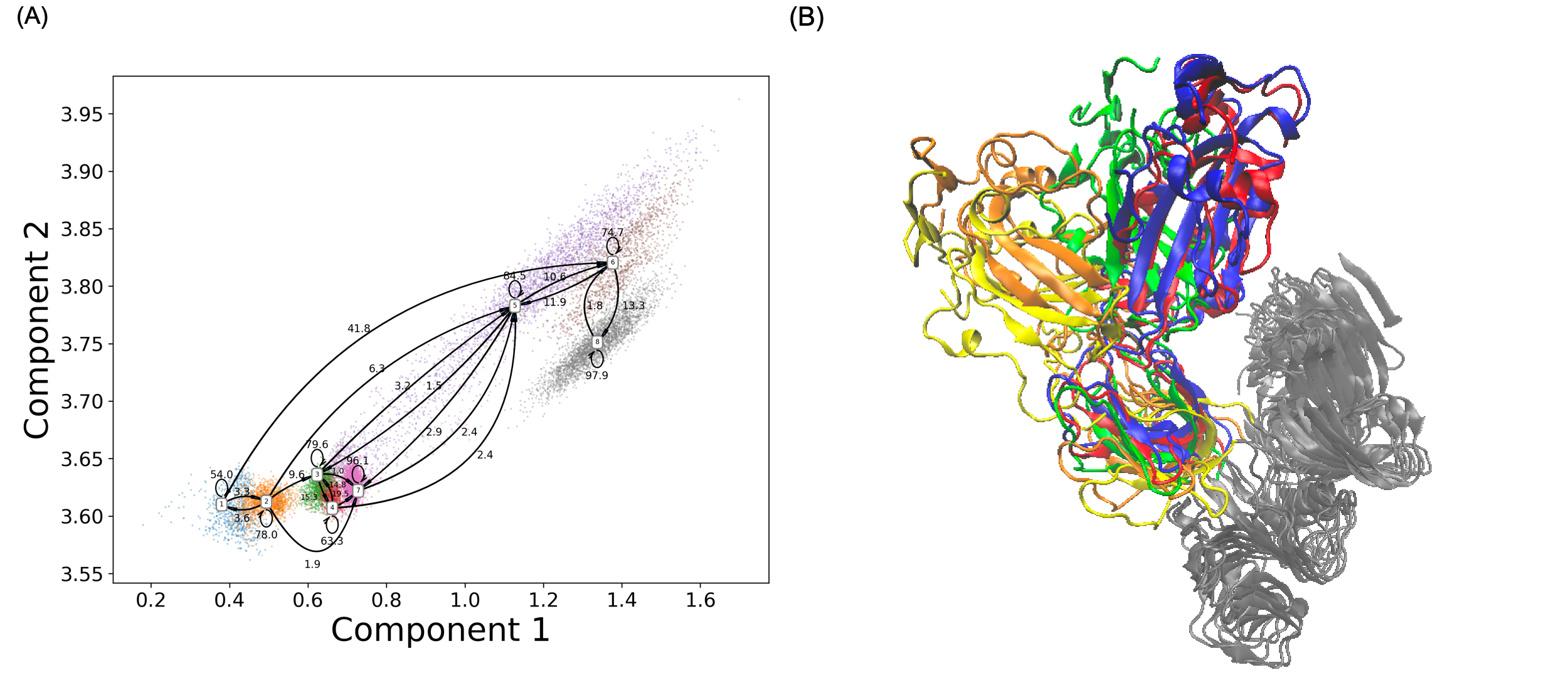
44. Tian, H.; Tao, P.* Deciphering the protein motion of S1 subunit in SARS-CoV-2 spike glycoprotein through integrated computational methods. 2020, Journal of Biomolecular Structure and Dynamics, 39, 6705 DOI: 10.1080/07391102.2020.1802338
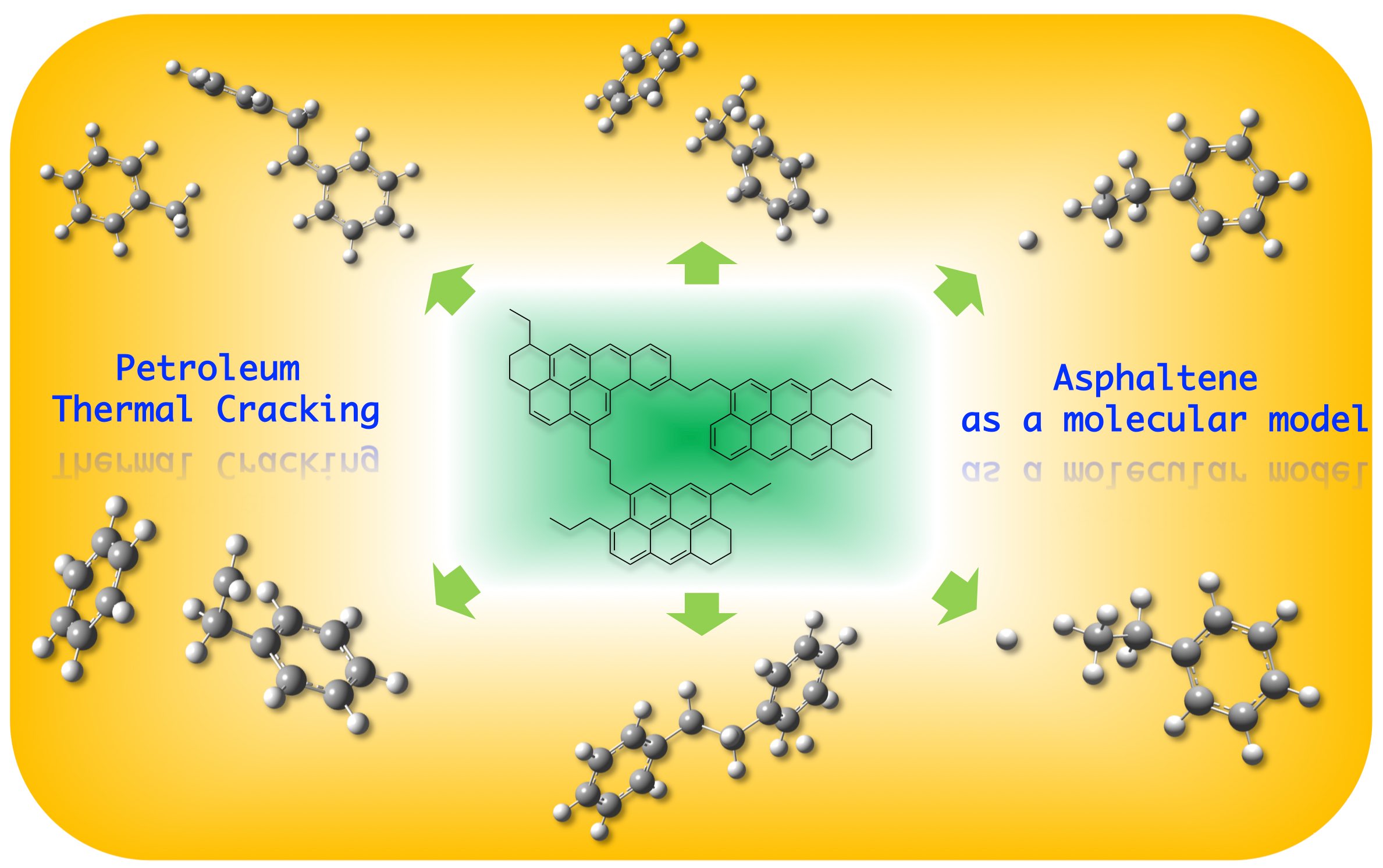
43. Wang, F.; Tao, P.* Exploring free energy profile of petroleum thermal cracking mechanisms. 2020, J. Mol. Mod., 26:15 DOI: 10.1007/s00894-019-4273-3
42. Wang, F.; Zhou, H.; Wang, X.; Tao, P.* Dynamical Behavior of β-Lactamases and Penicillin-Binding Proteins in Different Functional States and Its Potential Role in Evolution. 2019, Entropy, 21(11), 1130 DOI: 10.3390/e21111130
41. Zhou, H.*; Wang, F.; Bennett, D. I. G.; Tao, P.* Directed kinetic transition network model. 2019, J. Chem. Phys. 151, 144112 DOI: 10.1063/1.5110896
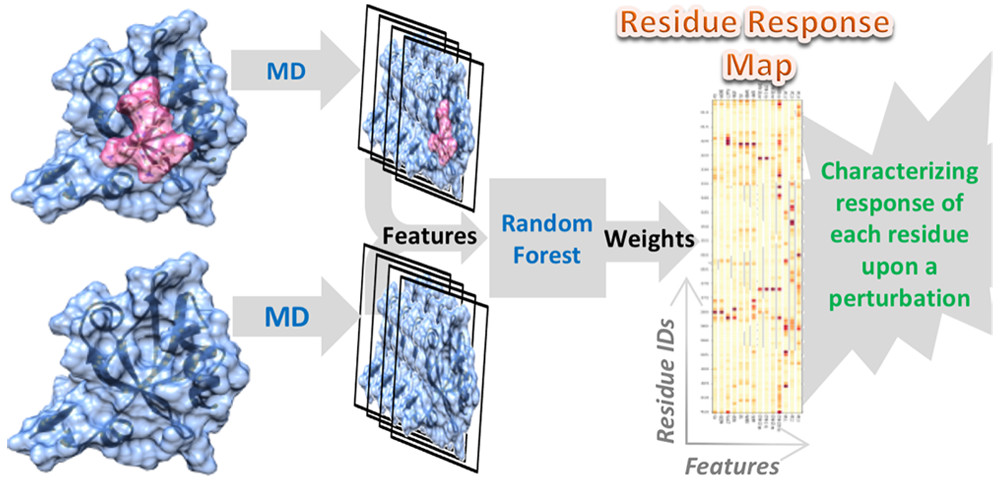
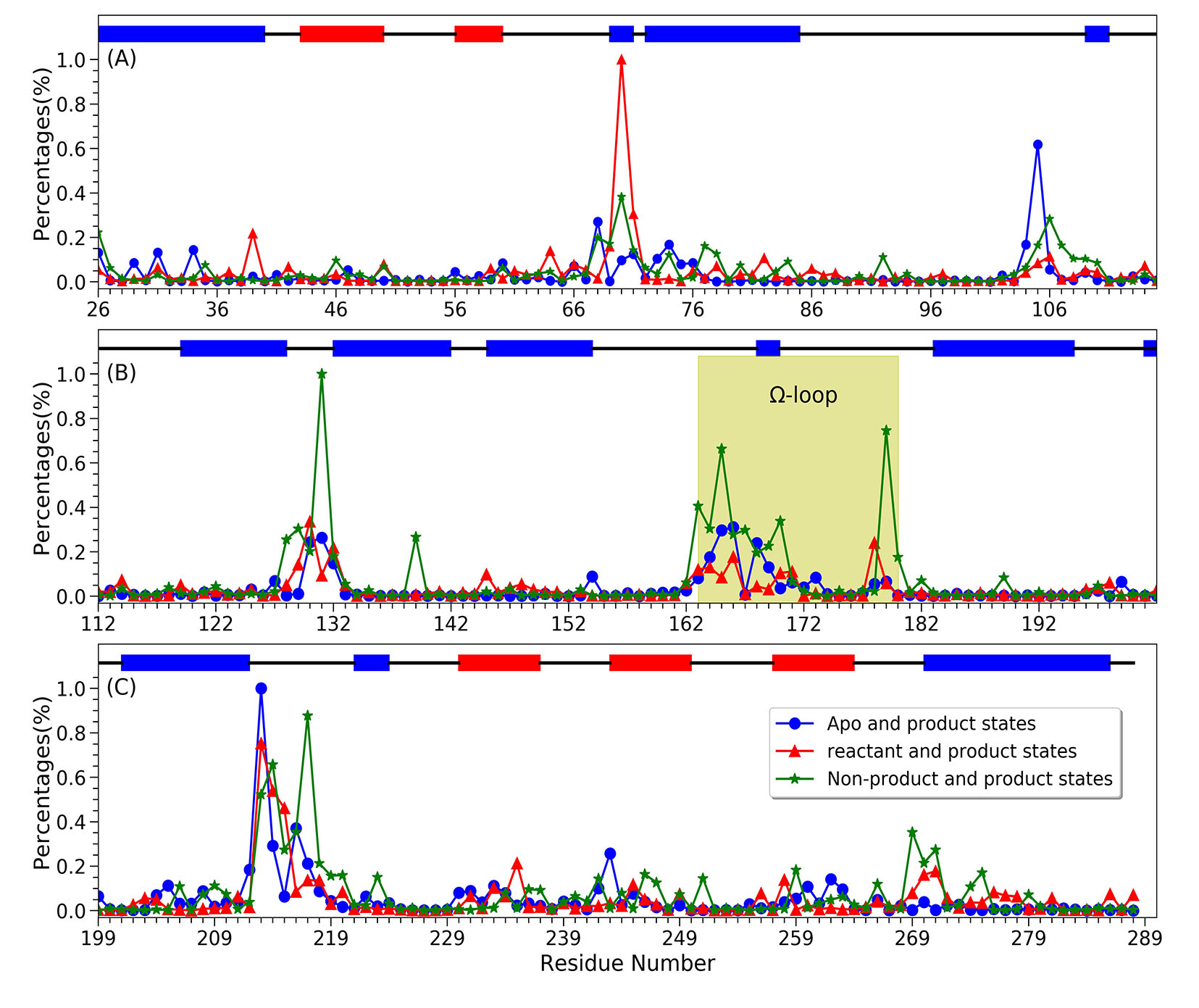
40. Hayatshahi, H. S.; Ahuactzin, E.; Tao, P.; Wang, S.; Liu, J.* Probing Protein Allostery as a Residue-Specific Concept via Residue Response Maps. 2019, J. Chem. Inf. Model. 59, 4691-4705 DOI: 10.1021/acs.jcim.9b00447
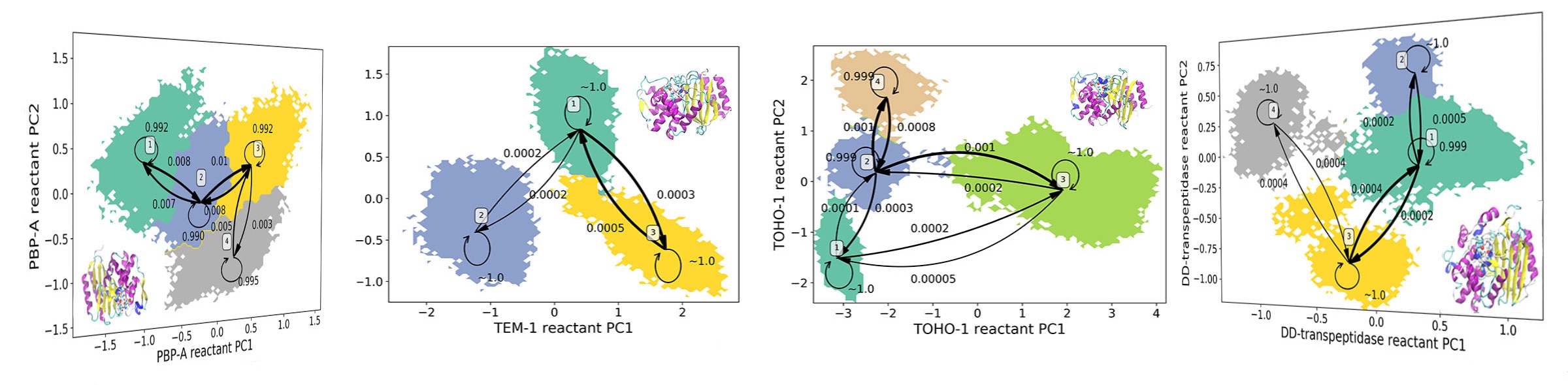
39. Wang, F.; Shen, L.; Zhou, H.; Wang, S.; Wang, X.; Tao, P.* Machine Learning Classification Model for Functional Binding Modes of TEM-1 β-Lactamase. 2019, Front. Mol. Biosci. 6:47 DOI: 10.3389/fmolb.2019.00047
38. Zhou, H.; Dong, Z.; Verkhivker, G.; Zoltowski, B. D.; Tao, P.*; Allosteric mechanism of the circadian protein Vivid resolved through Markov state model and machine learning analysis. PLoS Comput Biol. 2019, 15(2): e1006801 DOI: 10.1371/journal.pcbi.1006801
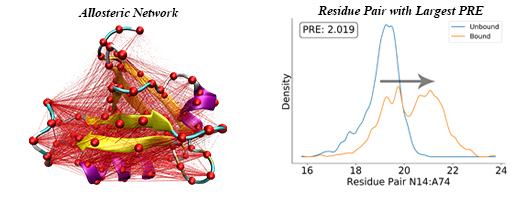
37. Zhou, H.; Tao, P.*; REDAN: relative entropy-based dynamical allosteric network model. Mol. Phys. 2018, 117, 1334. DOI: 10.1080/00268976.2018.1543904
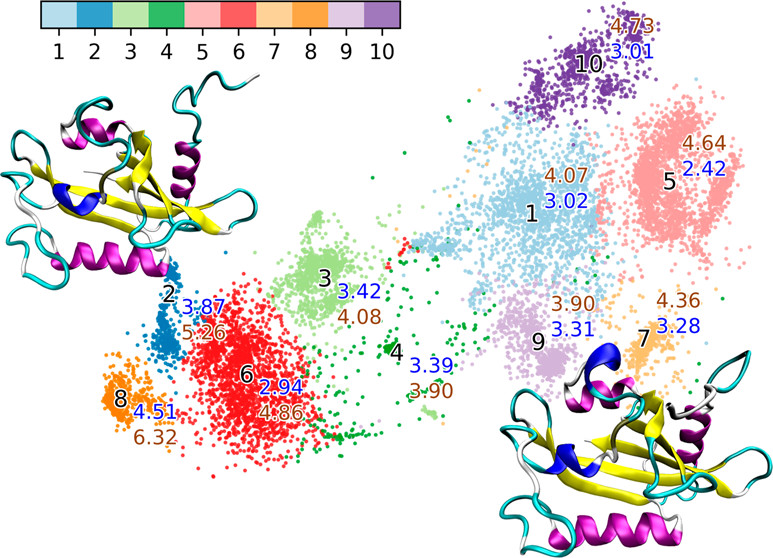
36. Zhou, H.; Wang, F.; Tao, P.*; t-Distributed Stochastic Neighbor Embedding (t-SNE) Method with the Least Information Loss for Macromolecular Simulations. J. Chem. Theory Comput. 2018, 14, 5499-5510. DOI: 10.1021/acs.jctc.8b00652
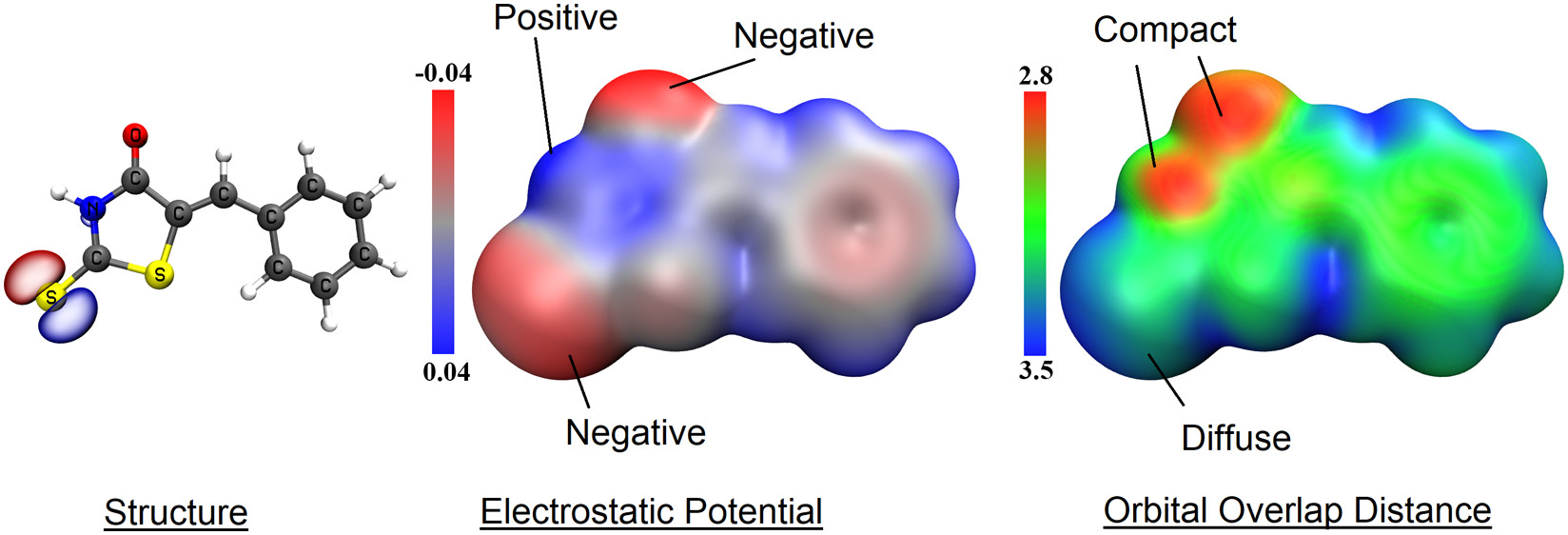
35. Mehmood, A.; Jones, S. I.; Tao, P.; Janesko, B. G.*, An Orbital-Overlap Complement to Ligand and Binding Site Electrostatic Potential Maps. J. Chem. Inf. Model. 2018, 58, 1836-1846. DOI: 10.1021/acs.jcim.8b00370
34. Zahler, C. T.; Zhou, H.; Abdolvdahabi, A.; Holden, R. L.; Rasouli, S.; Tao, P.; Shaw, B. F. Direct Measurement of Charge Regulation in Metalloprotein ElectronTransfer, Angew. Chem. Int. Ed. 2018, 57, 5364-5368 DOI: 10.1002/anie.201712306
This paper was selected as Very Important Paper (VIP) by the journal editor and featured on the journal cover.
33. Zhou, H.; Dong, Z.; Tao, P.*; Novel Application of Machine Learning Approaches on the Recognition of Protein States and Identification of Function-Related Residues. J. Comput. Chem., 2018, 39, 1481-1490 DOI: 10.1002/jcc.25218
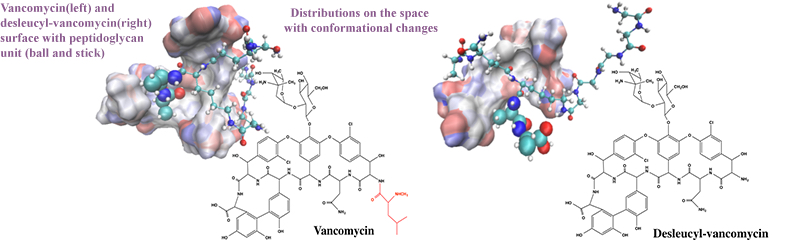
32. Wang, F.; Zhou, H.; Olademehin, O. P.; Kim, S. J.; Tao, P. Insights into Key Interactions between Vancomycin and Bacterial Wall Structures. ACS Omega 2018, 3, 37−45 DOI: 10.1021/acsomega.7b01483
Check out a short online liveslides presentation featuring this work. link
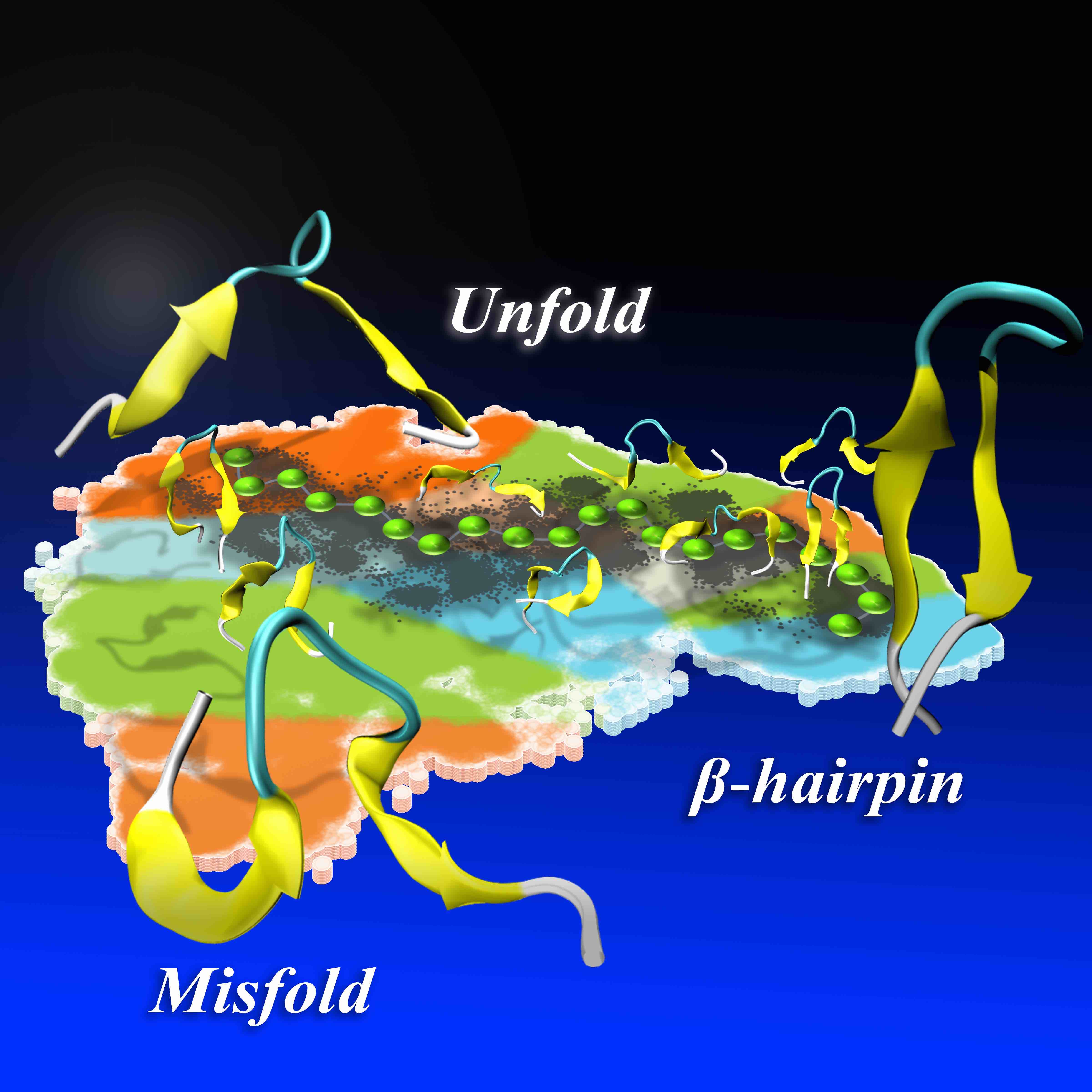
31. Zhou, H.; Tao, P. Dynamics Sampling in Transition Pathway Space. J. Chem. Theory Comput. 2018, 14, 14−29 DOI: 10.1021/acs.jctc.7b00606
Check out a short online liveslides presentation featuring this work. link
30. Dong, Z.; Zhou, H.; Tao, P. Combining protein sequence, structure, and dynamics: A novel approach for functional evolution analysis of PAS domain superfamily. Protein Science 2017, 27, 421-430 DOI: 10.1002/pro.3329
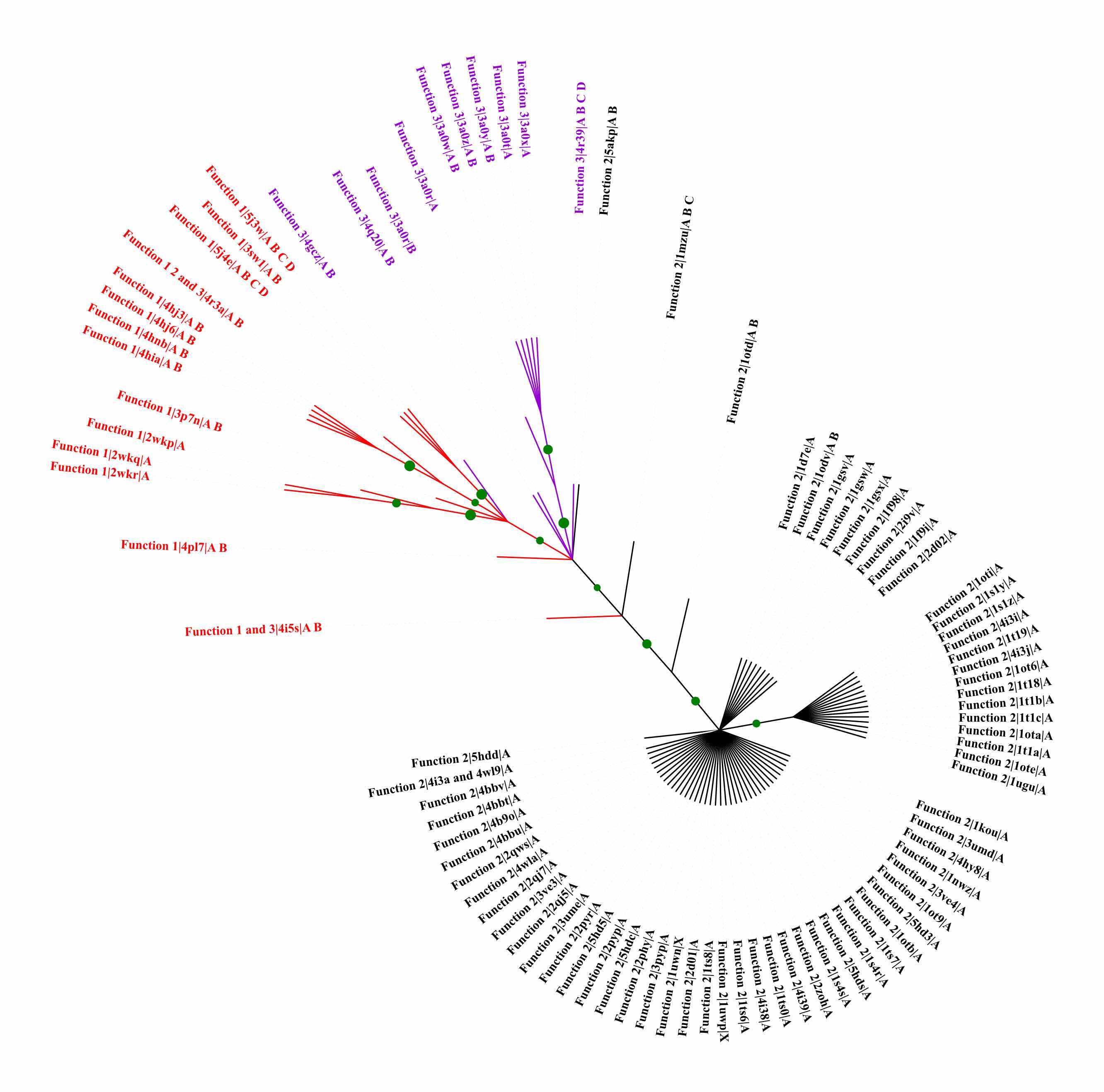
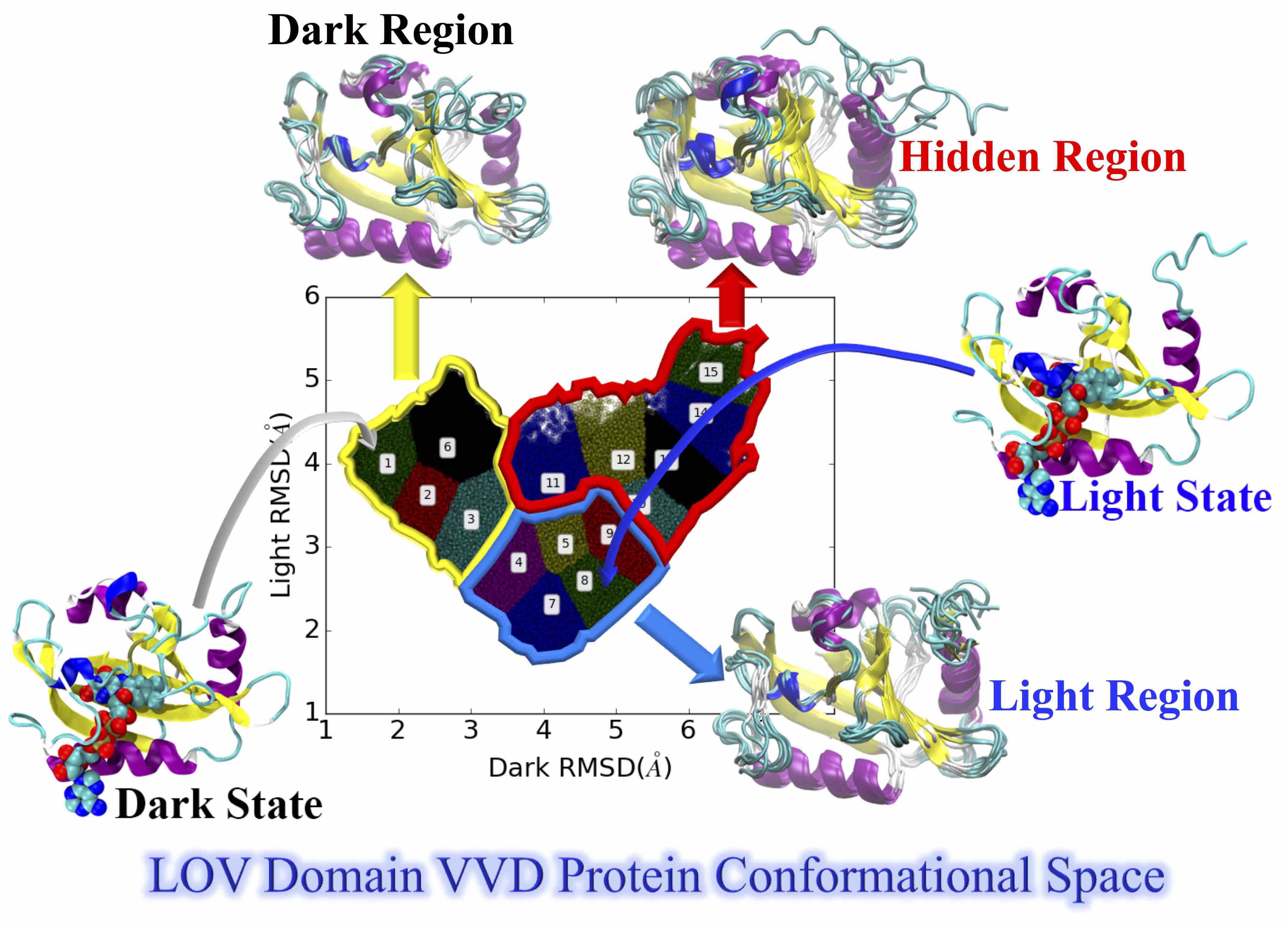
29. Zhou, H.; Zoltowski, B. D.; Tao, P. Revealing Hidden Conformational Space of LOV Protein VIVID Through Rigid Residue Scan Simulations. Scientific Reports 2017, 7, 46626 DOI: 10.1038/srep46626
28. Tao, P.; Hackett, J. C.; Kim, J. Y.; Saffen, D.; Hayes, C. J.; Hadad, C. “Molecular Determinants of TRPC6 Channel Recognition by FKBP12” in Computational Chemistry Methodology in Structural Biology and Materials Sciences, ISBN 978-1-77188-568-3, Apple Academic Press, Inc., 2017.
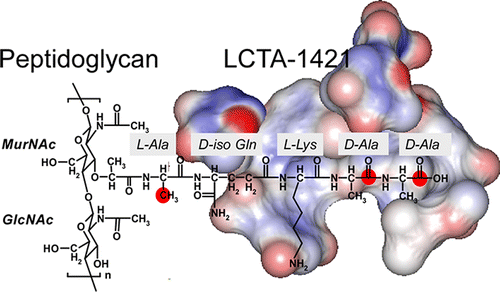
27. Chang, J.; Zhou, H.; Preobrazhenskaya, M.; Tao, P.;Kim, S. J. The Carboxyl Terminus of Eremomycin Facilitates Binding to the Non-d-Ala-d-Ala Segment of the Peptidoglycan Pentapeptide Stem. Biochemistry 2016, 55, 3383–3391. DOI: 10.1021/acs.biochem.6b00188
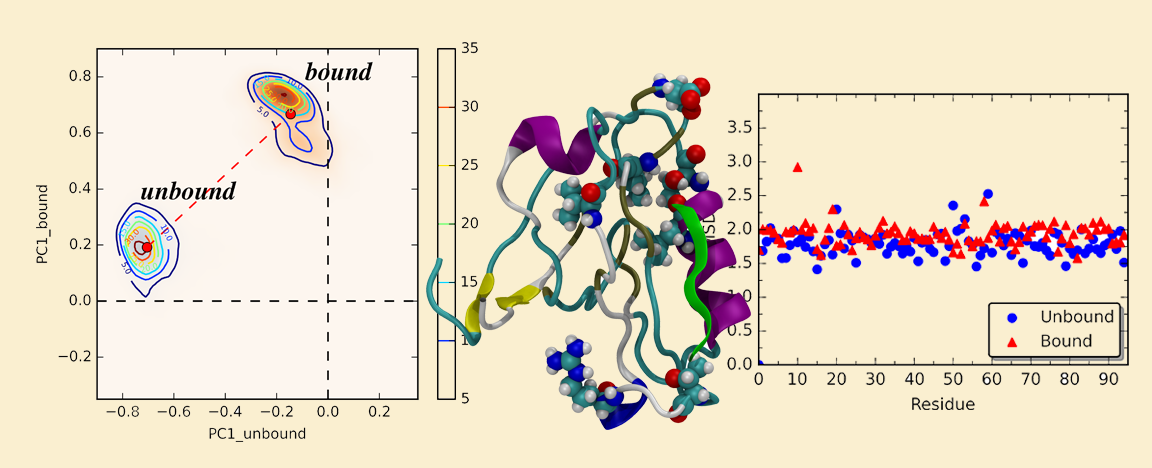
26. Kalescky, R.; Zhou, H.; Liu, J.; Tao, P. (2016) Rigid Residue Scan Simulations Systematically Reveal Residue Entropic Roles in Protein Allostery. PLoS Comput Biol 12(4): e1004893. DOI: 10.1371/ journal.pcbi.1004893
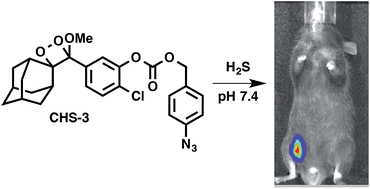
25. Cao, J.; Lopez, R.; Thacker J. M.; Moon, J. Y.; Jiang, C.; Morris, S. N. S.; Bauer, J. H.; Tao, P.; Mason, R. P.; Lippert, A. R.* "Chemiluminescent Probes for Imaging H2S in Living Animals." Chem. Sci. 2015, 6, 1979-1985. DOI: 10.1039/C4SC03516J
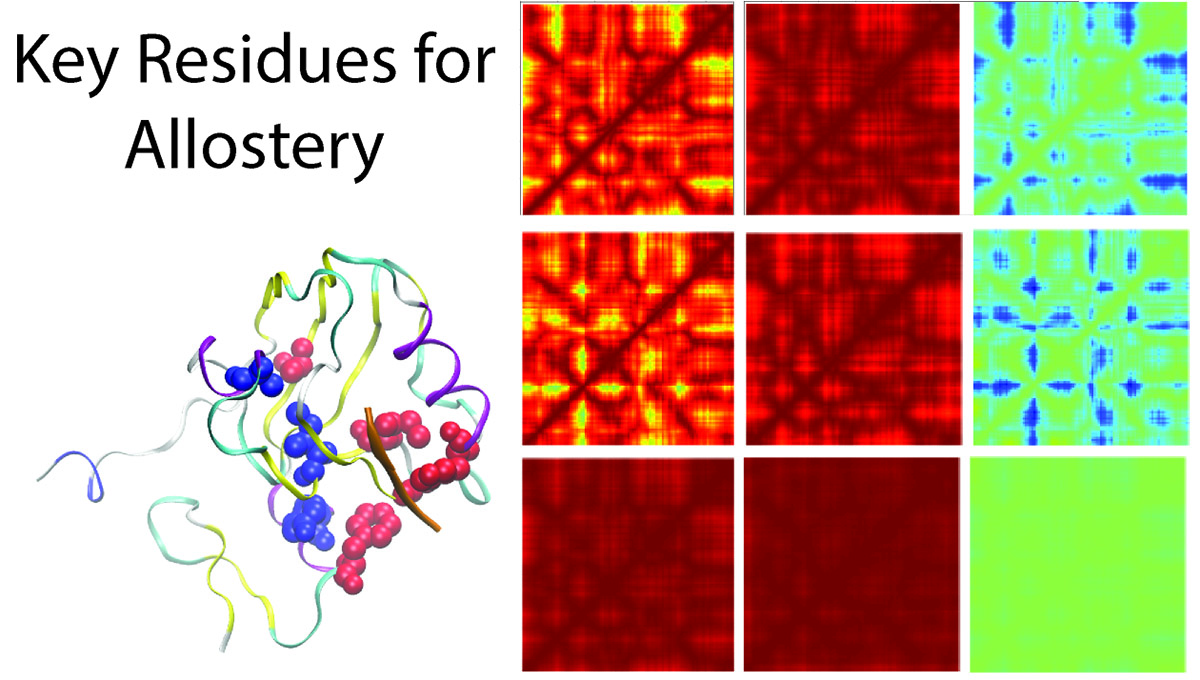
24. Kalesky, R.; Liu, J.; Tao, P.; “Identifying Key Residues for Protein Allostery through Rigid Residue Scan”. J. Phys. Chem. A 2015, 119, 1689-1700. DOI: 10.1021/jp5083455
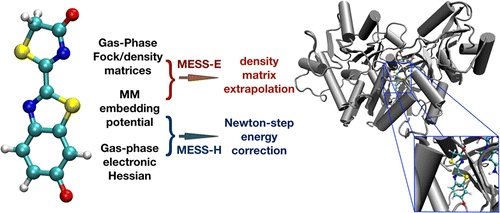
23. Sodt, A. J.; Mei, Y.; König, G.; Tao, P.; Steele, R.; Brooks, B. R.; Shao, Y. “Multiple Environment Single System Quantum Mechanical/Molecular Mechanical (MESS-QM/MM) Calculations. 1. Estimation of Polarization Energies”. J. Phys. Chem. A 2015, 119, 1511-1523. DOI: 10.1021/jp5072296
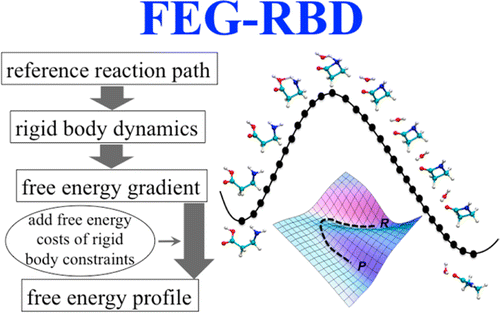
22. Tao, P.; Alexander J. Sodt, Yihan Shao, Gerhard König, Bernard R. Brooks; “Computing the Free Energy along a Reaction Coordinate Using Rigid Body Dynamics”. J. Chem. Theory Comput. 2014, 10, 4198-4207. DOI: 10.1021/ct500342h
Publications Unaffiliated with SMU:
21. Tao, P.; Larkin, J. D.; Brooks, B. R. “Reaction Path Optimization and Sampling Methods and Their Applications for Rare Events” in Some Applications of Quantum Mechanics, ISBN 979-953-51-0059-1, InTech, 2012. DOI: 10.5772/35351
20. Tao, P.; Hodošček, M.; Larkin, J. D; Shao, Y.; Brooks, B. R; “Comparison of Three Chain-of-States Methods: Nudged Elastic Band, Replica Path with Restraints or Constraints”. J. Chem. Theory Comput. 2012, 8, 5035-5051. DOI: 10.1021/ct3006248
19. Tao, P.; Wu, X.; Brooks, B. R; “Maintain Rigid Structures in Verlet Based Cartesian Molecular Dynamics Simulations”. J. Chem. Phys. 2012, 137, 134110. DOI:10.1063/1.4756796
18. Tao, P.; Parquette, J. R; Hadad, C. M; “Right- and Left-Handed Helices, What is in between? Interconversion of Helical Structures of Alternating Pyridinedicarboxamide/m-(phenylazo)azobenzene Oligomers”. J. Chem. Theory Comput. 2012, 8, 5137-5149. DOI:10.1021/ct2009335
17. Bao, X.; Tao, P.; Villamena, F. A.; Hadad, C. M; “Spin Trapping of Hydroperoxyl Radical by a Cyclic Nitrone Conjugated to β-Cyclodextrin: A Computational Study”. Theor. Chem. Acc. 2012, 131, 1248-1257. DOI: 10.1007/s00214-012-1248-1
16. Zhang, S.; Qu, Z.; Tao, P.; Brooks, B. R; Shao, Y.; Chen, X.; Liu, C.; "Quantum Chemical Study of the Ground and Excited State Electronic Structures of Carbazole Oligomers with and without Triarylborane Substitutes", J. Phys. Chem. C, 2012, 116, 12434-12442. DOI: 10.1021/jp3027447
15. Zhou, J.; Tao, P.; Fisher, J. F.; Shi, Q.; Mobashery, S.; Schlegel, H. B. “QM/MM Studies of the Matrix Metalloproteinase 2 (MMP2) Inhibition Mechanism of (S) SB-3CT and its Oxirane Analogue”, J. Chem. Theory Comput. 2010, 6, 3580-3587. (Selected as cover article) DOI: 10.1021/ct100382k
14. Tao, P.; Gatti, D. L.; Schlegel, H. B. “Common Basis for the Mechanism of Metallo and Non-metallo KDO8P Synthases”, J. Inorg. Biochem. 2010, 104, 1267-1275. DOI: 10.1016/j.jinorgbio.2010.08.008
13. Psciuk, T. B.; Tao, P.; Schlegel, H. B. “Ab Initio Classical Trajectory Study of the Fragmentation of C3H4 Dications on the Singlet and Triplet Surfaces”, J. Phys. Chem. A 2010, 114, 7653-7660. DOI: 10.1021/jp102238g
12. Tao, P.; Schlegel, H. B. “A Toolkit to Assist ONIOM Calculations”, J. Comput. Chem. 2010, 31, 2363-2369. DOI: 10.1002/jcc.21524
11. Sanan, T. T.; Muthukrishnan, S.; Beck, J. M.; Tao, P.; Hayes, C. J.; Otto, T. C.; Cerasoli, D. M.; Lenz, D. E.; Hadad, C. M. “Computational modeling of human paraoxonase 1: preparation of protein models, binding studies, and mechanistic insights”, J. Phys. Org. Chem., 2010, 23, 357-369. DOI: 10.1002/poc.1678
10. Tao, P.; Fisher, J. F.; Shi, Q.; Mobashery, S.; Schlegel, H. B. “Matrix Metalloproteinase 2 (MMP2) Inhibition: DFT and QM/MM Studies of the Deprotonation-Initialized Ring-Opening Reaction of Sulfoxide Analog of SB-3CT”, J. Phys. Chem. B, 2010, 114, 1030-1037. DOI: 10.1021/jp909327y
9. Tao, P.; Gatti, D. L.; Schlegel, H. B. “The energy landscape of 3-Deoxy- D-manno-octulosonate 8-Phosphate Synthase”, Biochemistry, 2009, 48, 11706-11714 DOI: 10.1021/bi901341h
8. Tao, P.; Fisher, J. F.; Shi, Q.; Vreven, T.; Mobashery, S.; Schlegel, H. B. “Matrix Metalloproteinase 2 (MMP2) Inhibition: Combined Quantum Mechanics and Molecular Mechanics Studies of the Inhibition Mechanism of (4-Phenoxyphenylsulfonyl) methylthiirane and Its Oxirane Analogue”, Biochemistry, 2009, 48, 9839-9847 DOI: 10.1021/bi901118r
7. Tao, P.; Fisher, J. F.; Mobashery, S.; Schlegel, H. B. “DFT Studies of the Ring-Opening Mechanism of SB-3CT, a Potent Inhibitor of Matrix Metalloproteinase 2”, Org. Lett., 2009, 11, 2559-2562 DOI: 10.1021/ol9008393
6. Kona, F.; Tao, P.; Martin, P.; Xu, X.; Gatti, D. L.. “Electronic Structure of the Metal Center in the Cd2+, Zn2+, and Cu2+ Substituted Forms of KDO8P Synthase: Implications for Catalysis”, Biochemistry, 2009, 48, 3610-3630 (Kona, K. and Tao, P. contributed equally to this work) DOI: 10.1021/bi801955h
5. King, E. D.; Tao, P.; Sanan, T. T.; Hadad, C. M.; Parquette, J. R. “Photomodulated chiral induction in helical azobenzene oligomers”. Org. Lett. 2008, 10, 1671-1674 DOI: 10.1021/ol8004722
4. Mendlik, M. T.; Tao, P.; Hadad, C. M.; Coleman, R. S.; Lowary, T. L. “Synthesis of L-Daunosamine and L-Ristosamine Glycosides via Photoinduced Aziridination. Conversion to Thioglycosides for Use in Glycosylation Reactions.” J. Org. Chem. 2006, 71, 8059-8070 DOI: 10.1021/jo061167z
3. Tao, P.; Lai, L. “Protein Ligand Docking Based on Empirical Method for Binding Affinity Estimation” J. Comput.-Aided Mol. Des. 2001, 15, 429-446. DOI: 10.1023/A:1011188704521
2. Tao, P.; Wang, R.; Lai, L. “Calculating Partition Coefficients of Peptides by the Addition Method” J. Mol. Mod. 1999, 5, 189-195. DOI: 10.1007/s008940050118
1. Tao, P.; Wang, R.; Lai, L. “Calculation of Peptide’s Partition Coefficients by Amino Acid Addition Method” Wuli Huaxue Xuebao 1999, 15, 449-453.